Learning Relations from Biomedical Corpora Using Dependency Tree

Learning Relations from Biomedical Corpora Using Dependency Tree Levels Sophia Katrenko, Pieter Adriaans Presentation: 2011. 21 임준호 (201160192)

Contents • • • Introduction Related Work Relation Learning Experiments Conclusion
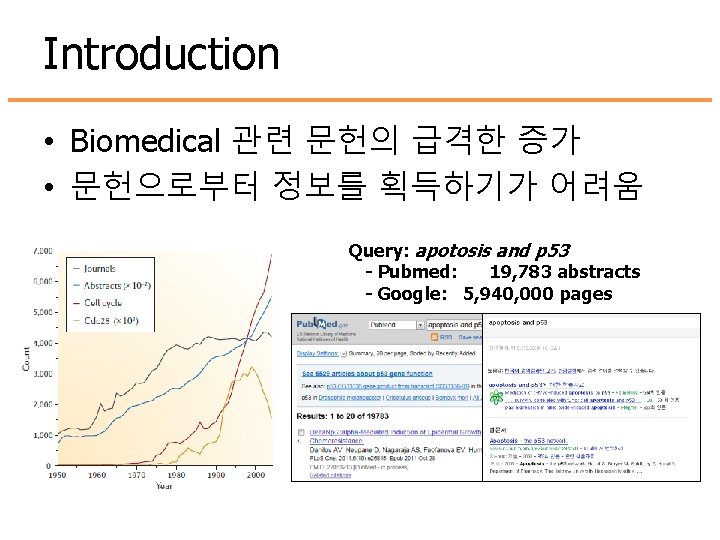
Introduction • Biomedical 관련 문헌의 급격한 증가 • 문헌으로부터 정보를 획득하기가 어려움 Query: apotosis and p 53 - Pubmed: 19, 783 abstracts - Google: 5, 940, 000 pages

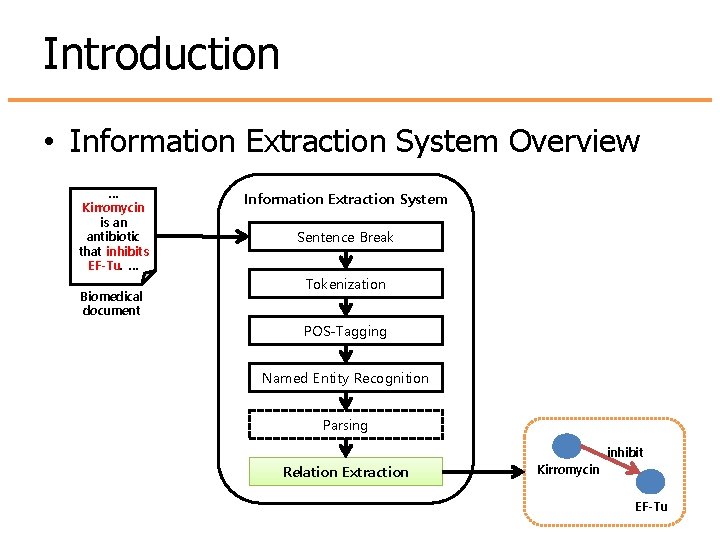
Introduction • Information Extraction System Overview … Kirromycin is an antibiotic that inhibits EF-Tu. … Biomedical document Information Extraction System Sentence Break Tokenization POS-Tagging Named Entity Recognition Parsing inhibit Relation Extraction Kirromycin EF-Tu
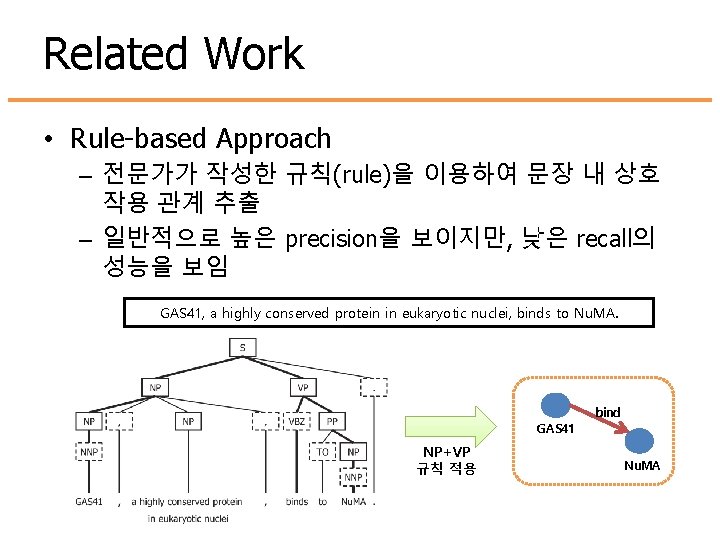
Related Work • Rule-based Approach – 전문가가 작성한 규칙(rule)을 이용하여 문장 내 상호 작용 관계 추출 – 일반적으로 높은 precision을 보이지만, 낮은 recall의 성능을 보임 GAS 41, a highly conserved protein in eukaryotic nuclei, binds to Nu. MA. bind GAS 41 NP+VP 규칙 적용 Nu. MA
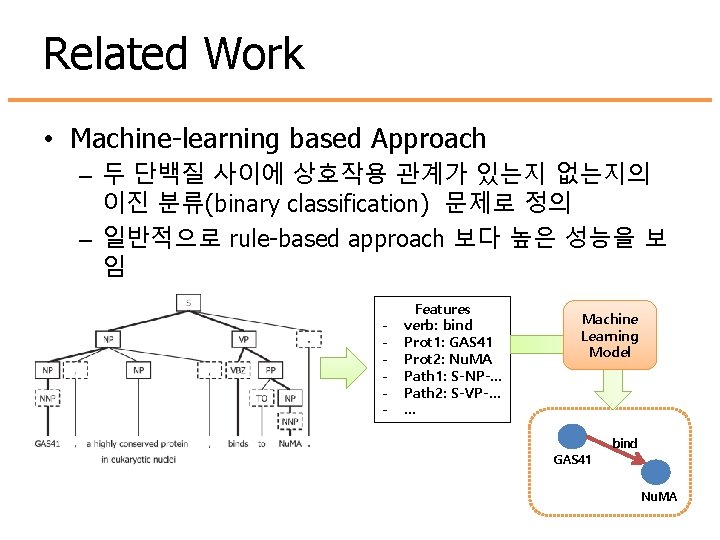
Related Work • Machine-learning based Approach – 두 단백질 사이에 상호작용 관계가 있는지 없는지의 이진 분류(binary classification) 문제로 정의 – 일반적으로 rule-based approach 보다 높은 성능을 보 임 - Features verb: bind Prot 1: GAS 41 Prot 2: Nu. MA Path 1: S-NP-… Path 2: S-VP-… … Machine Learning Model bind GAS 41 Nu. MA
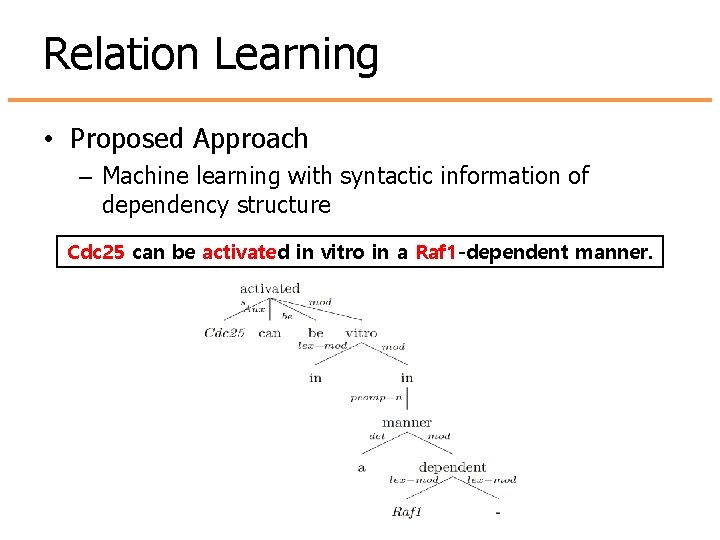
Relation Learning • Proposed Approach – Machine learning with syntactic information of dependency structure Cdc 25 can be activated in vitro in a Raf 1 -dependent manner.
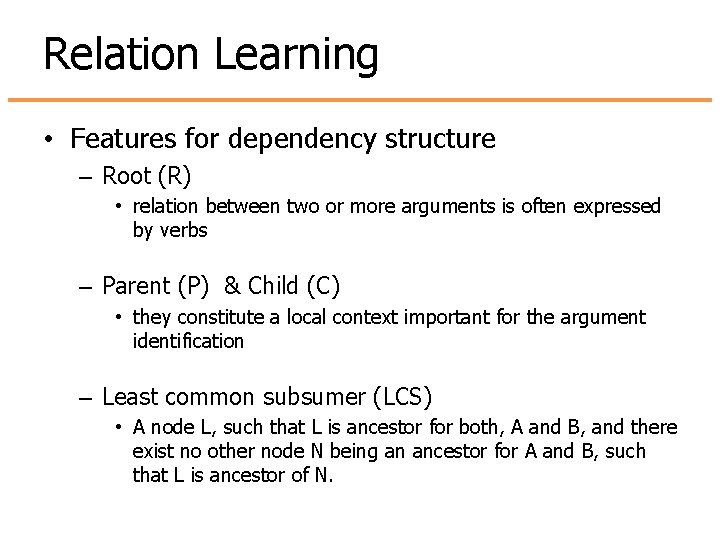
Relation Learning • Features for dependency structure – Root (R) • relation between two or more arguments is often expressed by verbs – Parent (P) & Child (C) • they constitute a local context important for the argument identification – Least common subsumer (LCS) • A node L, such that L is ancestor for both, A and B, and there exist no other node N being an ancestor for A and B, such that L is ancestor of N.
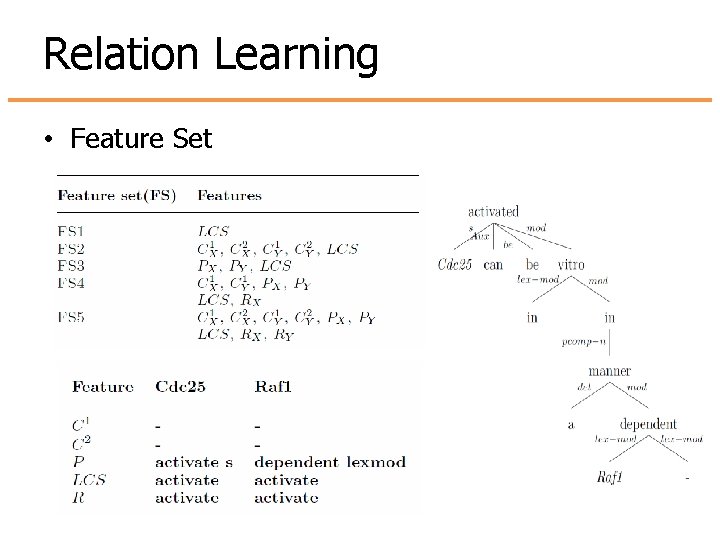
Relation Learning • Feature Set
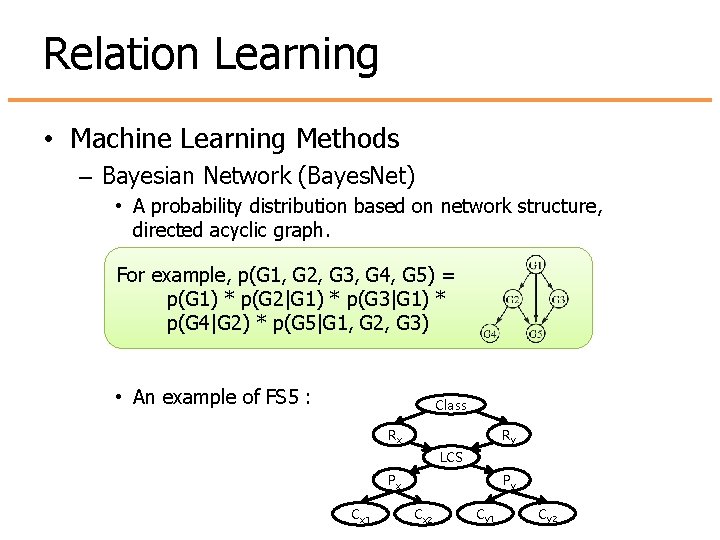
Relation Learning • Machine Learning Methods – Bayesian Network (Bayes. Net) • A probability distribution based on network structure, directed acyclic graph. For example, p(G 1, G 2, G 3, G 4, G 5) = p(G 1) * p(G 2|G 1) * p(G 3|G 1) * p(G 4|G 2) * p(G 5|G 1, G 2, G 3) • An example of FS 5 : Class Rx Ry LCS Px Cx 1 Py Cx 2 Cy 1 Cy 2
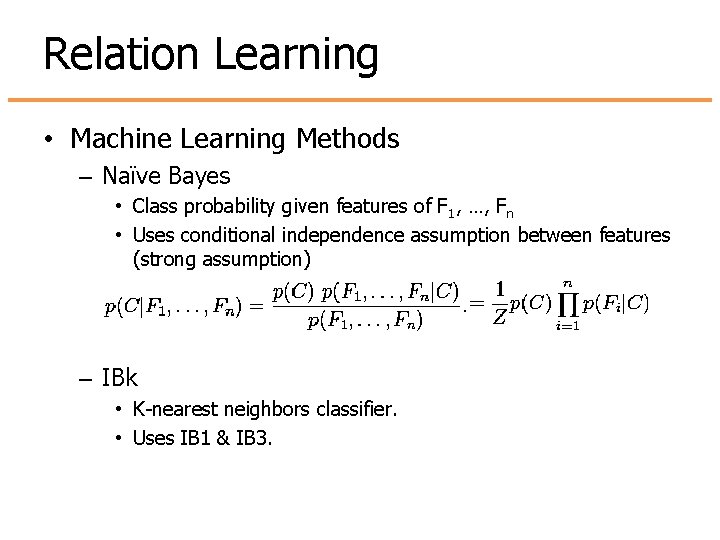
Relation Learning • Machine Learning Methods – Naïve Bayes • Class probability given features of F 1, …, Fn • Uses conditional independence assumption between features (strong assumption) – IBk • K-nearest neighbors classifier. • Uses IB 1 & IB 3.
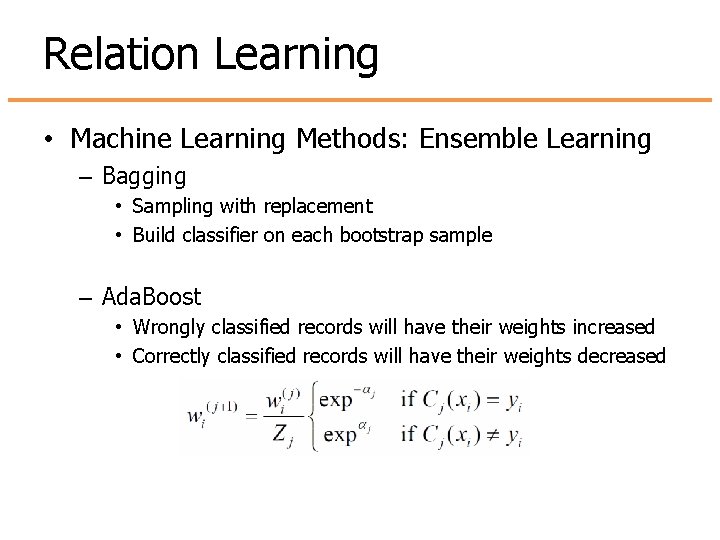
Relation Learning • Machine Learning Methods: Ensemble Learning – Bagging • Sampling with replacement • Build classifier on each bootstrap sample – Ada. Boost • Wrongly classified records will have their weights increased • Correctly classified records will have their weights decreased
Relation Learning • Machine Learning Methods: Ensemble Learning – Stacking • Used to combine machine learning models of different types • Procedures 1. 2. 3. 4. Split the training set into two disjoint sets. Train several base learners on the first part. Test the base learners on the second part. Using the predictions from 3) as the inputs, and the correct responses as the outputs, train a higher level learner. • Meta-learner combines the base learners, possibly nonlinearly.
Experiments • Experimental Environment – Corpora • AImed corpus – Created for PPI extraction method comparison – 225 Pubemd abstracts – 4, 736 training instances (985 of them are positive) • LLL corpus – Shared dataset for the Learning Language in Logic 2005 – 873 training instances (161 of them are positive) – Use WEKA machine learning toolkit
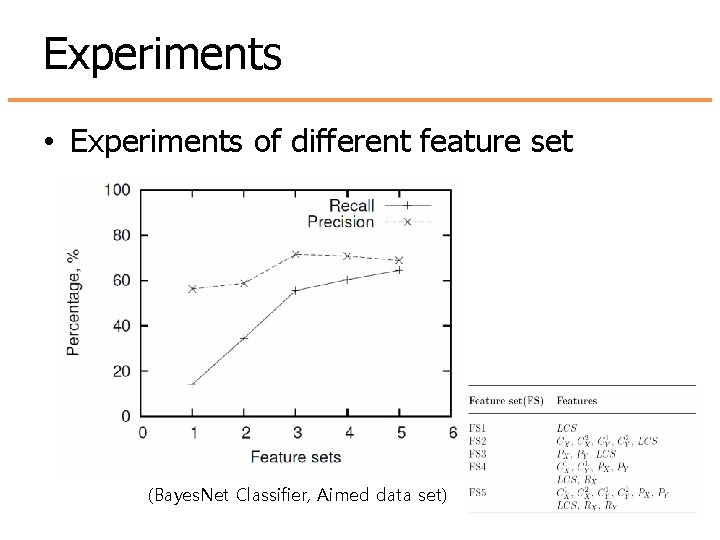
Experiments • Experiments of different feature set (Bayes. Net Classifier, Aimed data set)
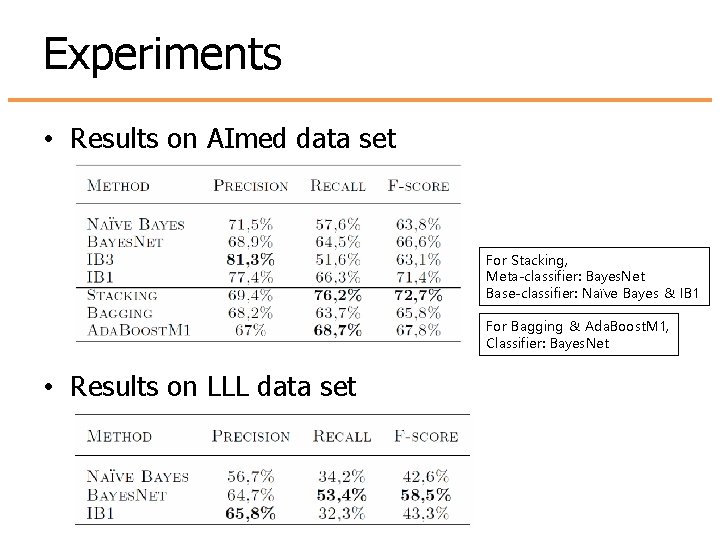
Experiments • Results on AImed data set For Stacking, Meta-classifier: Bayes. Net Base-classifier: Naïve Bayes & IB 1 For Bagging & Ada. Boost. M 1, Classifier: Bayes. Net • Results on LLL data set
Conclusion • Propose a relation learning method based on the dependency trees – Extracts gene-protein interaction (LLL) – Extracts protein-protein interaction (AImed) • Get better performance than the previous work of Bunescu and Mooney (2005)
- Slides: 18