Lateral organization and electrostatic control of signaling Proteinmembrane
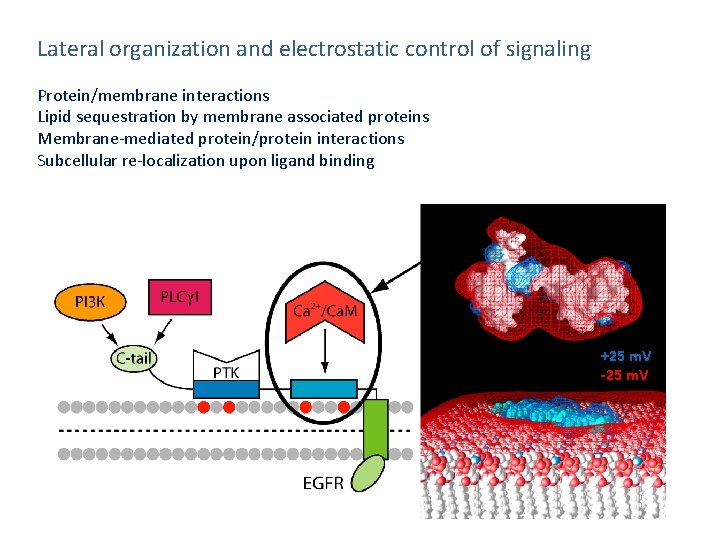
Lateral organization and electrostatic control of signaling Protein/membrane interactions Lipid sequestration by membrane associated proteins Membrane-mediated protein/protein interactions Subcellular re-localization upon ligand binding +25 m. V -25 m. V
High-throughput modeling of cellular signaling proteins Protein families involved in membrane-mediated subcellular targeting Within and across families, within and across genomes → Developed methods to reliably model all instances of peripheral proteins Tonya Silkov: Nebojsa Mirkovic: Hunjoong Lee: Frank Indiviglio: Paul Murray: Anna Mulgrew-Nesbitt: Janey Li: Zheng Li: Meng Wang: Mariam Konate: Arabidopsis 2010 project Phosphoinositide-binding domains in the human genome Structural genomics high-throughput modeling Database structures for integrating structures and predicited models Formation of retroviruses and host/virion interactions Phosphoinositide-modifying enzymes: PTEN and PLCs Synaptic vesicle exocytosis Simulation of the structure and dynamics of phosphoinositides Integration of structure-focused tools into ge. Workbench Modeling of phosphoinositide signaling http: //ph 7 wnu. cpmc. columbia. edu/
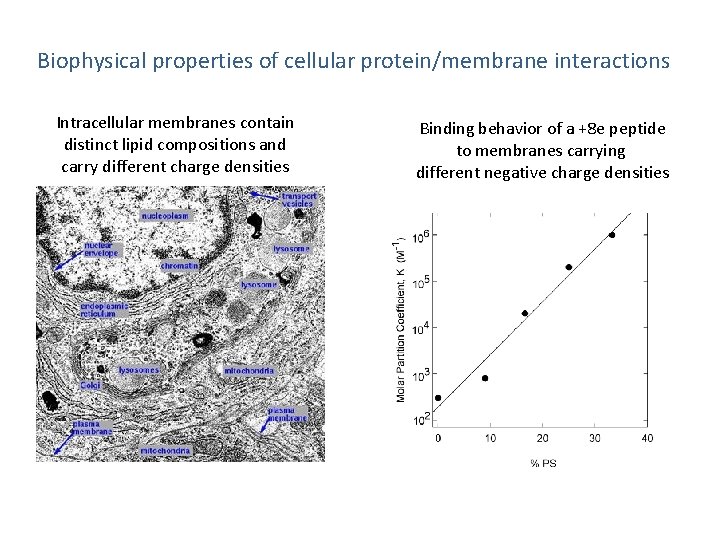
Biophysical properties of cellular protein/membrane interactions Intracellular membranes contain distinct lipid compositions and carry different charge densities Binding behavior of a +8 e peptide to membranes carrying different negative charge densities
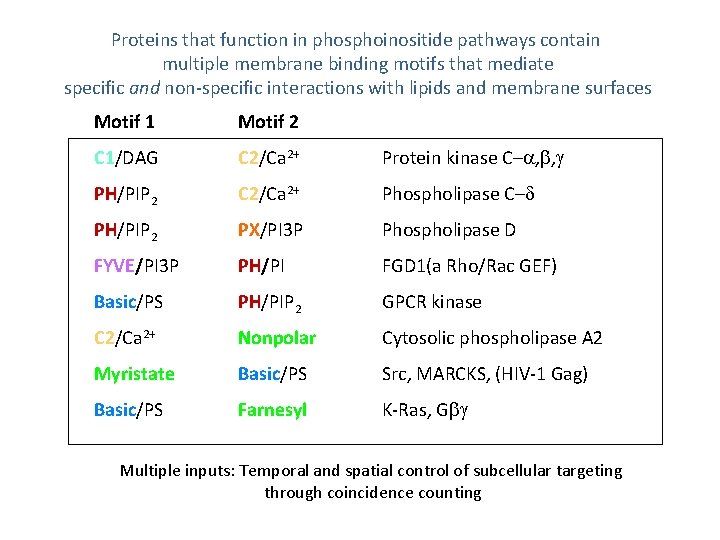
Proteins that function in phosphoinositide pathways contain multiple membrane binding motifs that mediate specific and non-specific interactions with lipids and membrane surfaces Motif 1 Motif 2 C 1/DAG C 2/Ca 2+ Protein kinase C– , , PH/PIP 2 C 2/Ca 2+ Phospholipase C– PH/PIP 2 PX/PI 3 P Phospholipase D FYVE/PI 3 P PH/PI FGD 1(a Rho/Rac GEF) Basic/PS PH/PIP 2 GPCR kinase C 2/Ca 2+ Nonpolar Cytosolic phospholipase A 2 Myristate Basic/PS Src, MARCKS, (HIV-1 Gag) Basic/PS Farnesyl K-Ras, G Multiple inputs: Temporal and spatial control of subcellular targeting through coincidence counting
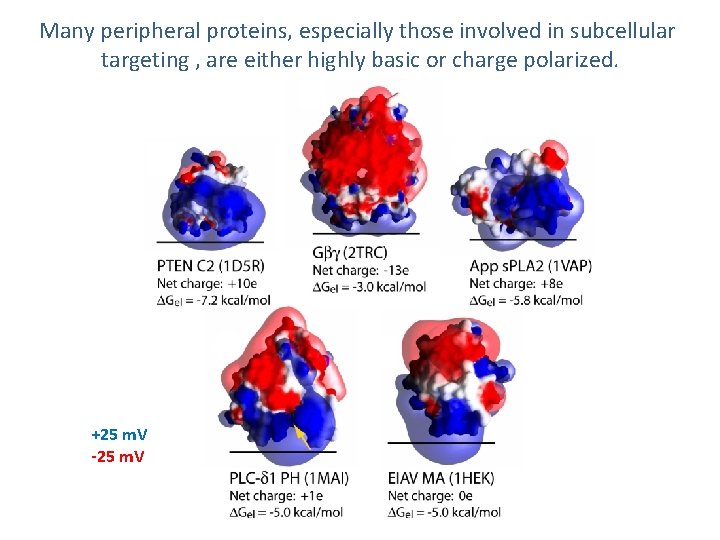
Many peripheral proteins, especially those involved in subcellular targeting , are either highly basic or charge polarized. +25 m. V -25 m. V
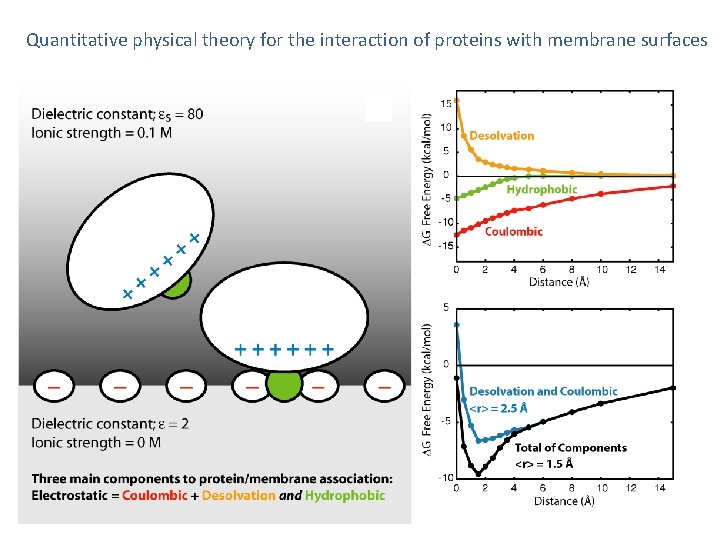
Quantitative physical theory for the interaction of proteins with membrane surfaces
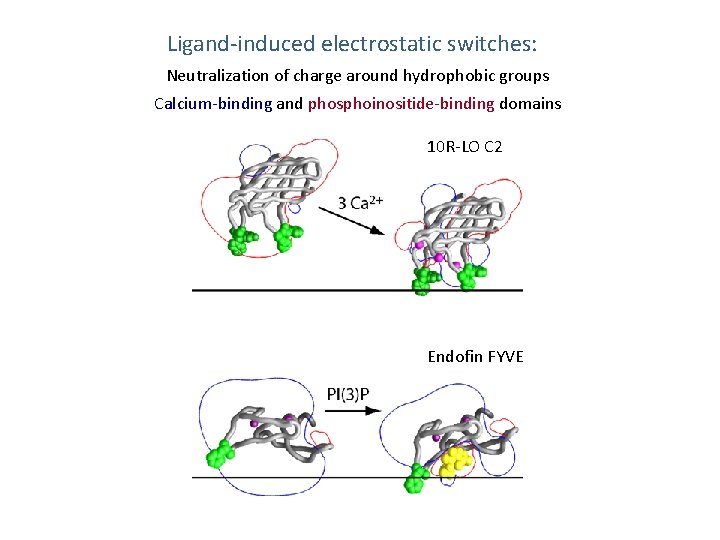
Ligand-induced electrostatic switches: Neutralization of charge around hydrophobic groups Calcium-binding and phosphoinositide-binding domains 10 R-LO C 2 Endofin FYVE
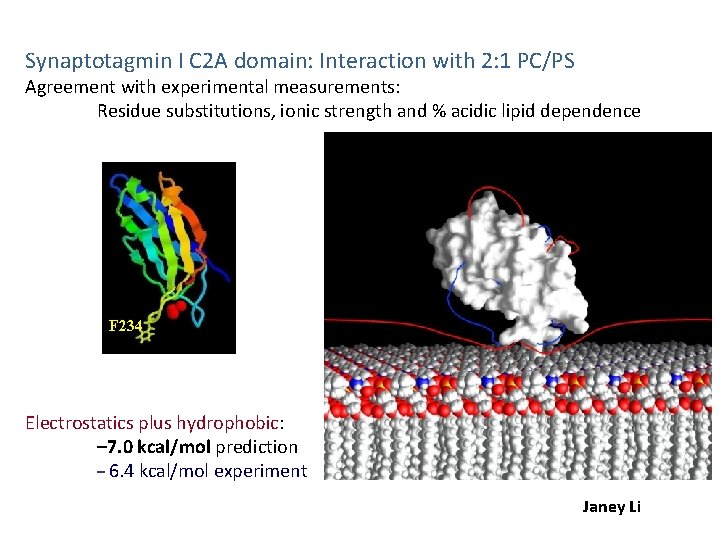
Synaptotagmin I C 2 A domain: Interaction with 2: 1 PC/PS Agreement with experimental measurements: Residue substitutions, ionic strength and % acidic lipid dependence F 234 Electrostatics plus hydrophobic: – 7. 0 kcal/mol prediction – 6. 4 kcal/mol experiment Janey Li

Connection among biophysical properties, membrane binding behavior, and subcellular localization No calcium Calcium Phospholipase C C 2 domains 5 -lipoxygenase C 2 domain Paul Murray
All lipid-binding domains in all model genomes Use what we have learned computationally and experimentally to develop: 1. More complete lists of peripheral proteins of known structure from the PDB; 2. Detect and model all instances of peripheral proteins in sequence databases; 3. Discover new instances, novel functionalities, new families; 4. Create databases to house this information; 5. Use this information to annotate protein sequences of unknown function.
Structural Genomics and Protein Family Analysis How to use computational tools to maximize the coverage of protein sequence/structure/function space “Leverage”: Number and quality of 3 D models produced from a set of structures as templates PSI 1 and PSI 2: NESG leverage ~220 sequence unique models NCBI non-redundant (nr) database: 6. 4 M protein sequences PDB Protein Data Bank: 0. 048 M protein structure → A judicious selection of structural targets with the modeling in hand, will provide almost complete coverage of protein sequence/structure space. There are many different ways to define “protein family” “Modelability”: Create “quality” models using known structures as templates Leverage for START domains: Modelability (7378) versus 30% sequence identity (2767)
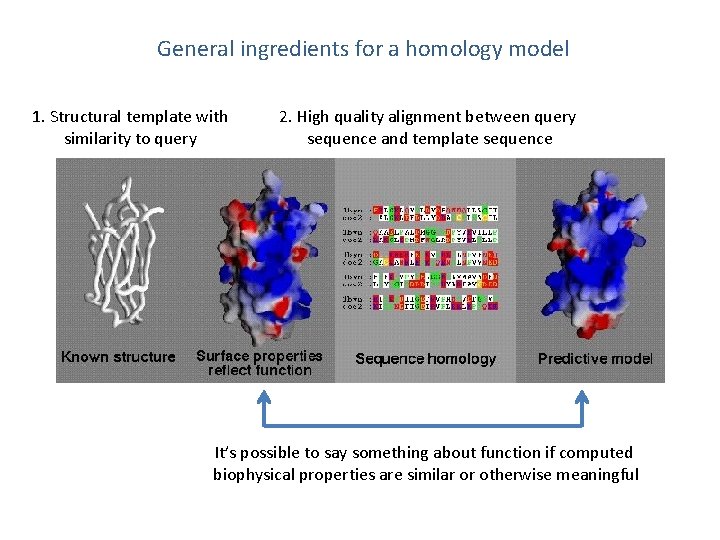
General ingredients for a homology model 1. Structural template with similarity to query 2. High quality alignment between query sequence and template sequence It’s possible to say something about function if computed biophysical properties are similar or otherwise meaningful
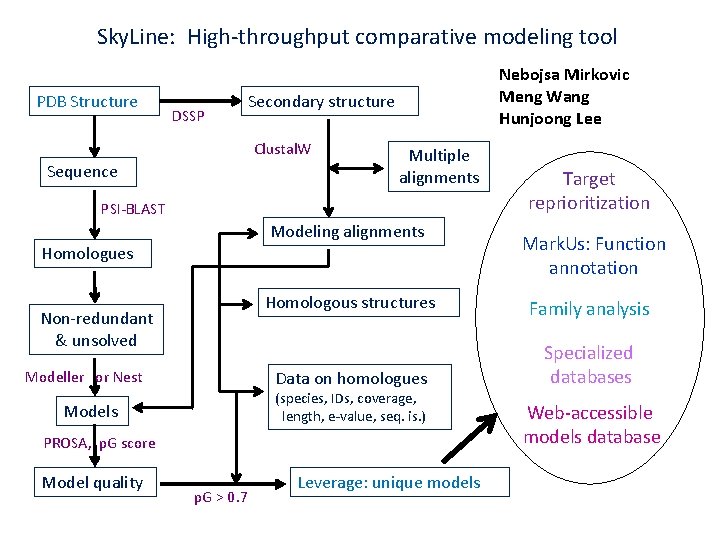
Sky. Line: High-throughput comparative modeling tool PDB Structure DSSP Secondary structure Clustal. W Sequence Nebojsa Mirkovic Meng Wang Hunjoong Lee Multiple alignments PSI-BLAST Modeling alignments Homologues Non-redundant & unsolved Modeller or Nest Family analysis Data on homologues Specialized databases PROSA, p. G score Model quality p. G > 0. 7 Mark. Us: Function annotation Homologous structures (species, IDs, coverage, length, e-value, seq. is. ) Models Target reprioritization Leverage: unique models Web-accessible models database
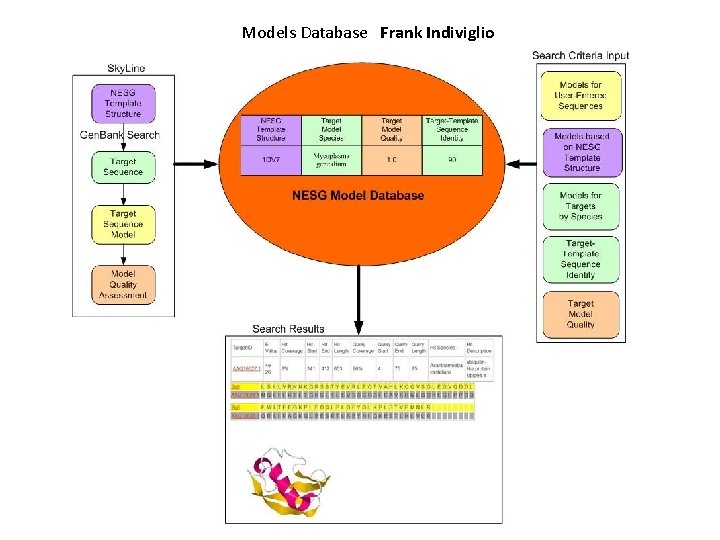
Models Database Frank Indiviglio
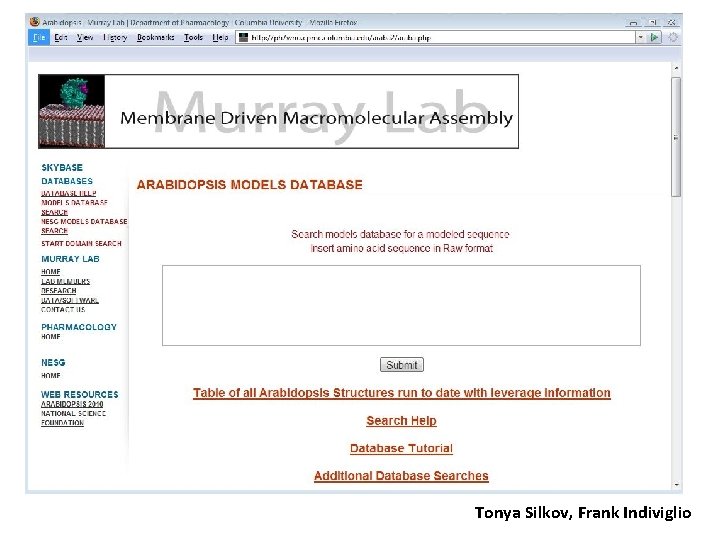
Tonya Silkov, Frank Indiviglio
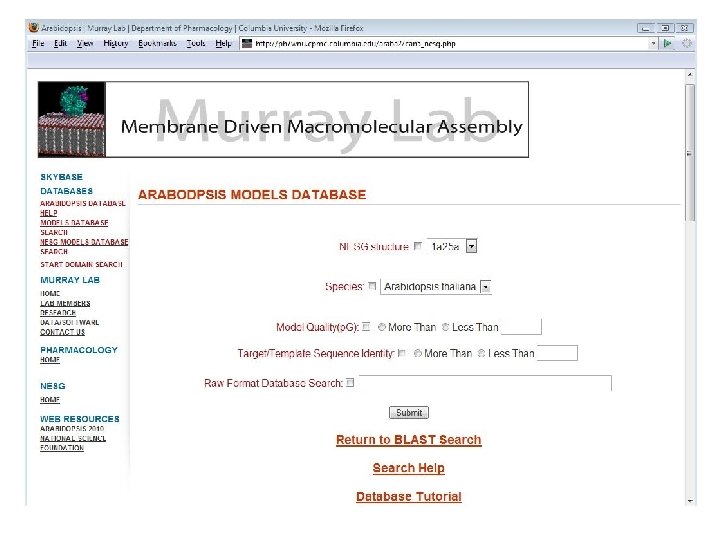
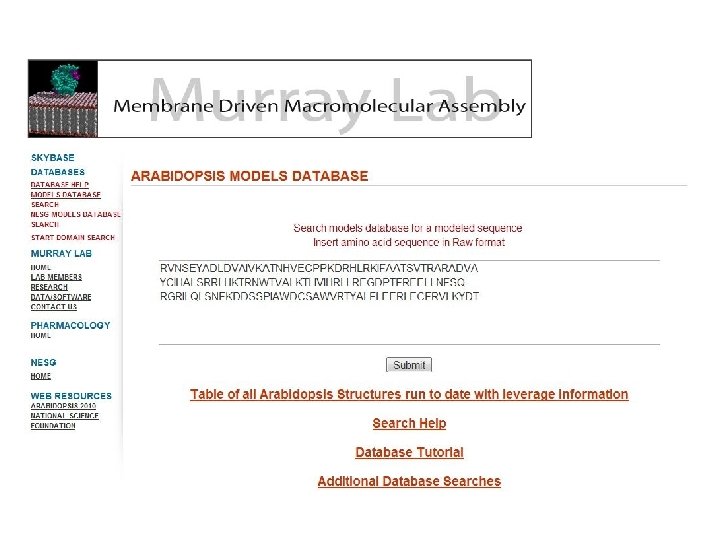
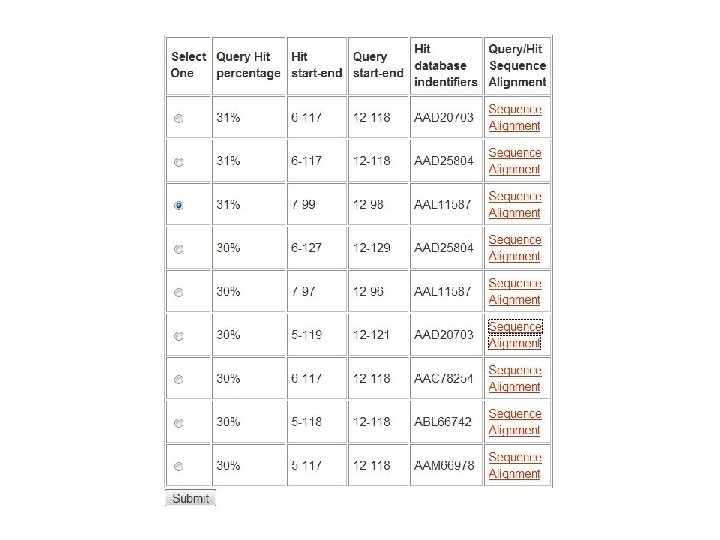
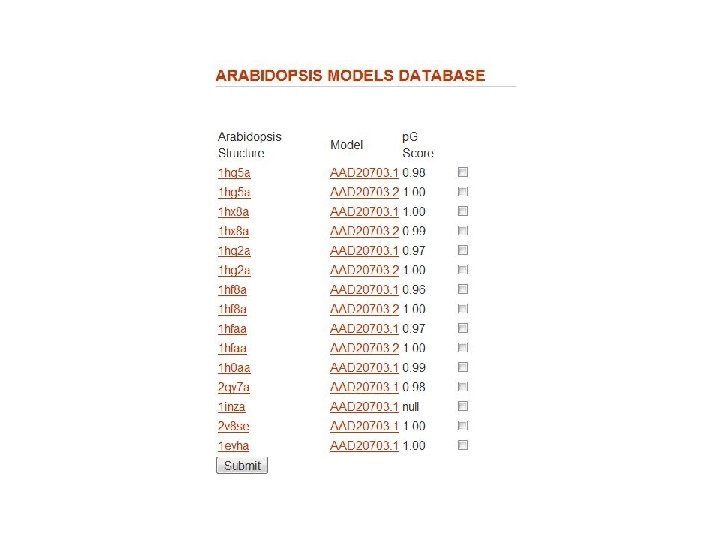
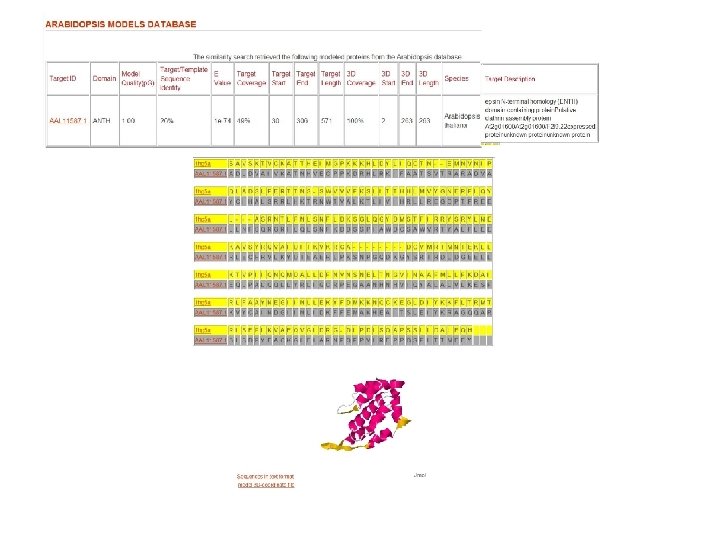
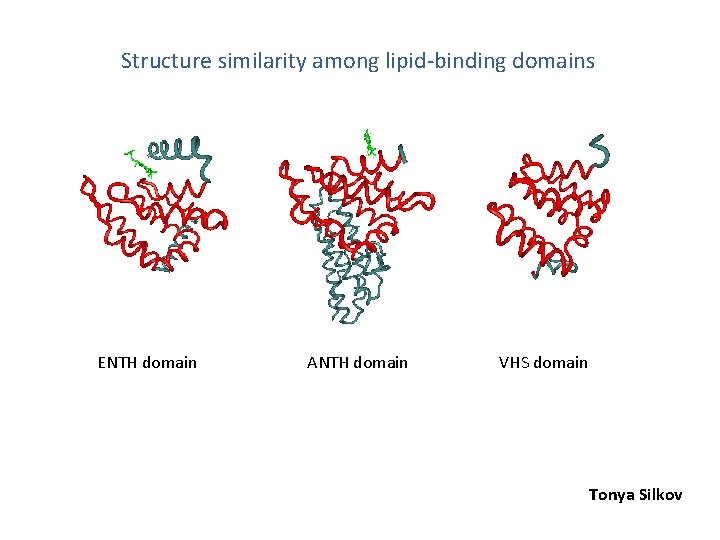
Fig. 1 Structure similarity among lipid-binding domains ENTH domain ANTH domain VHS domain Tonya Silkov
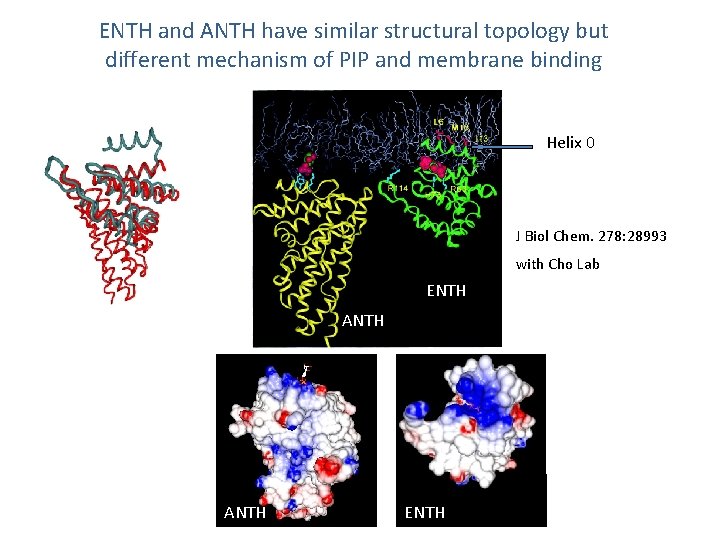
ENTH and ANTH have similar structural topology but different mechanism of PIP and membrane binding Helix 0 J Biol Chem. 278: 28993 with Cho Lab ENTH ANTH ENTH
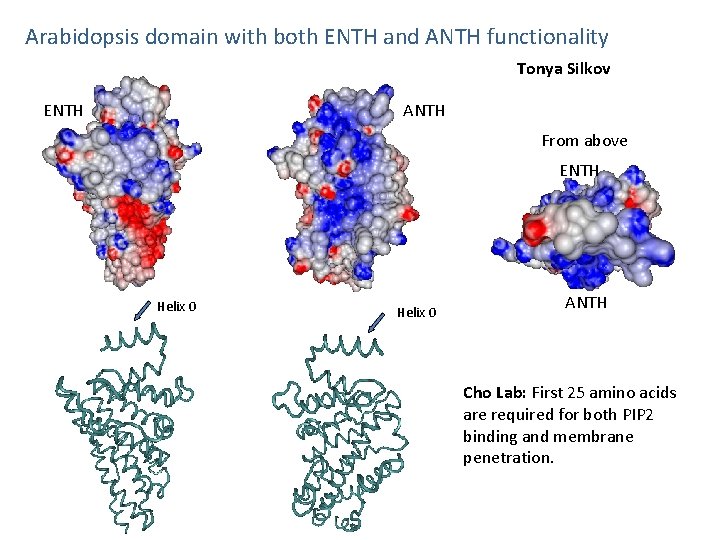
Arabidopsis domain with both ENTH and ANTH functionality Tonya Silkov ENTH ANTH From above ENTH Helix 0 ANTH Cho Lab: First 25 amino acids are required for both PIP 2 binding and membrane penetration.
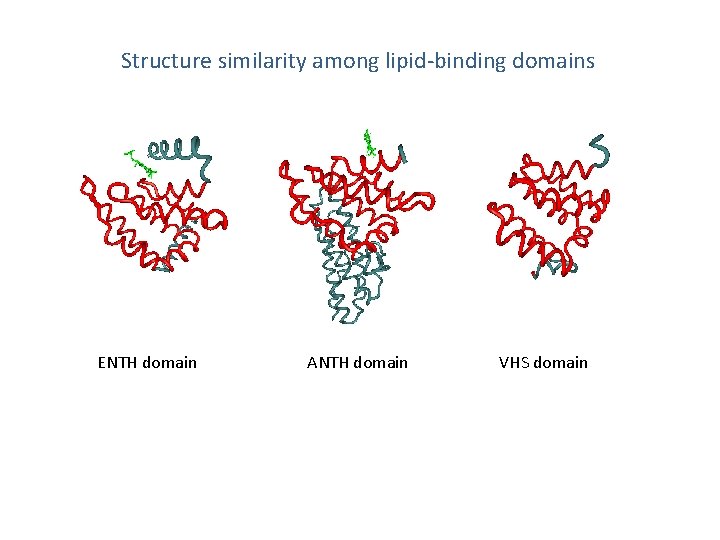
Fig. 1 Structure similarity among lipid-binding domains ENTH domain ANTH domain VHS domain
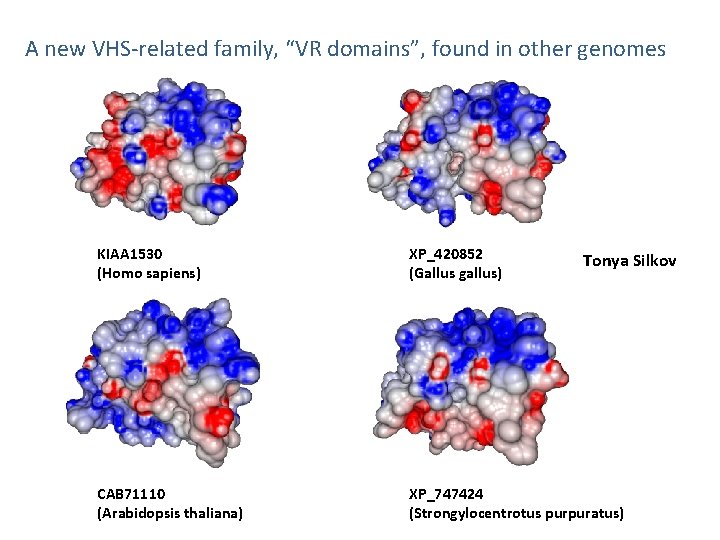
A new VHS-related family, “VR domains”, found in other genomes KIAA 1530 (Homo sapiens) XP_420852 (Gallus gallus) CAB 71110 (Arabidopsis thaliana) XP_747424 (Strongylocentrotus purpuratus) Tonya Silkov
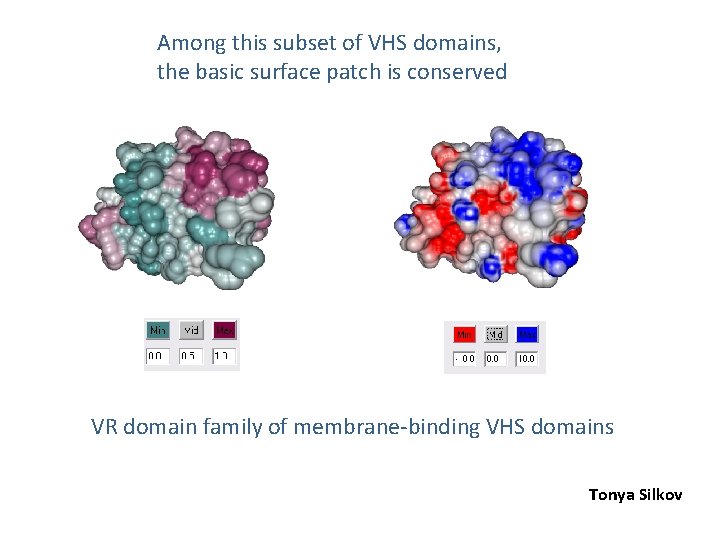
Among this subset of VHS domains, the basic surface patch is conserved VR domain family of membrane-binding VHS domains Tonya Silkov
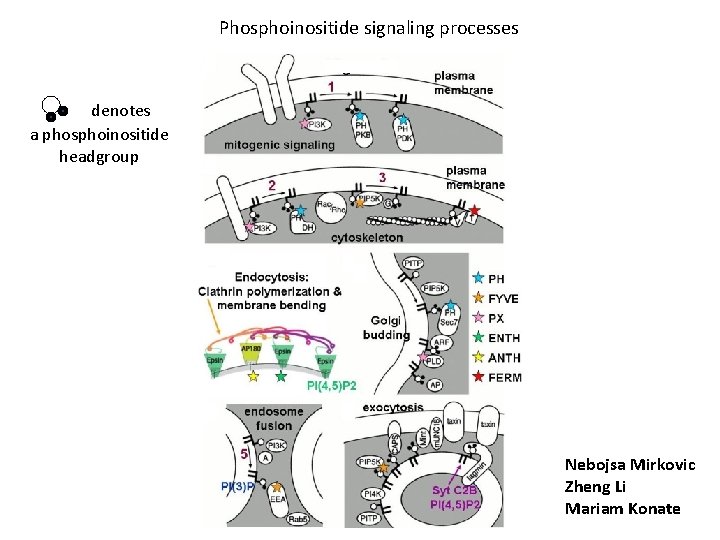
Phosphoinositide signaling processes denotes a phosphoinositide headgroup Nebojsa Mirkovic Zheng Li Mariam Konate

The ability to construct a quality model of a sequence is a more strategic definition of a protein family member Allows for the discovery of distantly related members With function annotation, allows for the discovery of new sub-groups Structures + Sequences -> Models + Function annotation (Markus) More comprehensive coverage of protein sequence/structure/function space
- Slides: 29