Laboratory of Molecular Genetics KNU Gene expression Analysis
Laboratory of Molecular Genetics, KNU Gene expression & Analysis
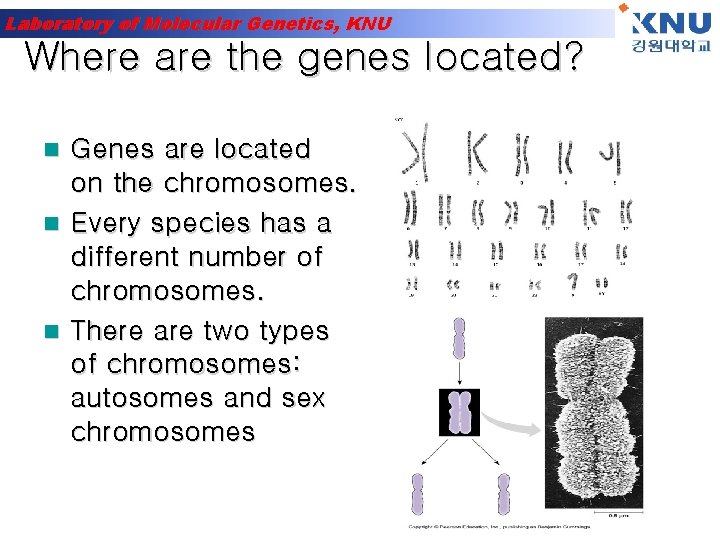
Laboratory of Molecular Genetics, KNU Where are the genes located? Genes are located on the chromosomes. n Every species has a different number of chromosomes. n There are two types of chromosomes: autosomes and sex chromosomes n
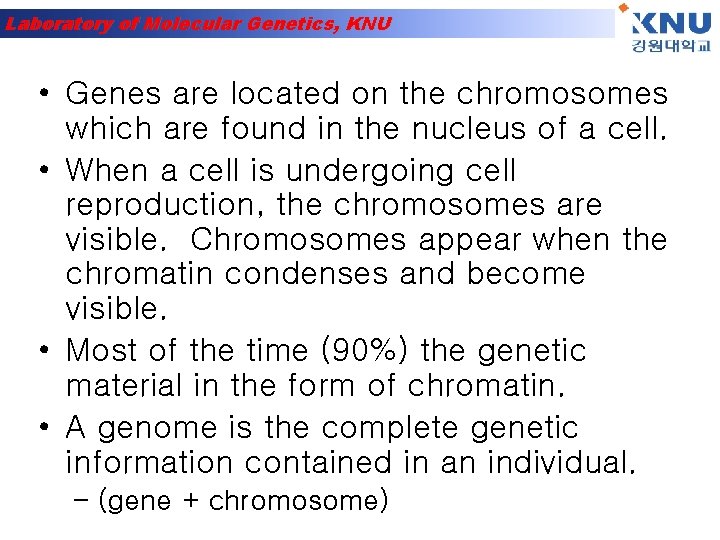
Laboratory of Molecular Genetics, KNU • Genes are located on the chromosomes which are found in the nucleus of a cell. • When a cell is undergoing cell reproduction, the chromosomes are visible. Chromosomes appear when the chromatin condenses and become visible. • Most of the time (90%) the genetic material in the form of chromatin. • A genome is the complete genetic information contained in an individual. – (gene + chromosome)
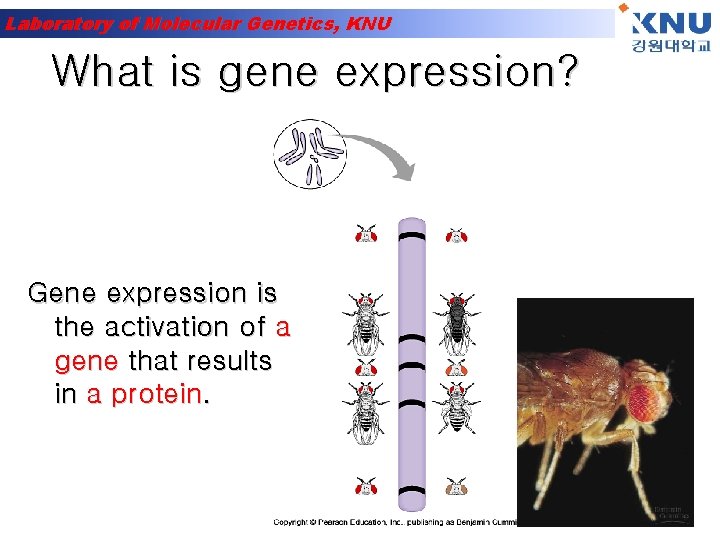
Laboratory of Molecular Genetics, KNU What is gene expression? Gene expression is the activation of a gene that results in a protein.
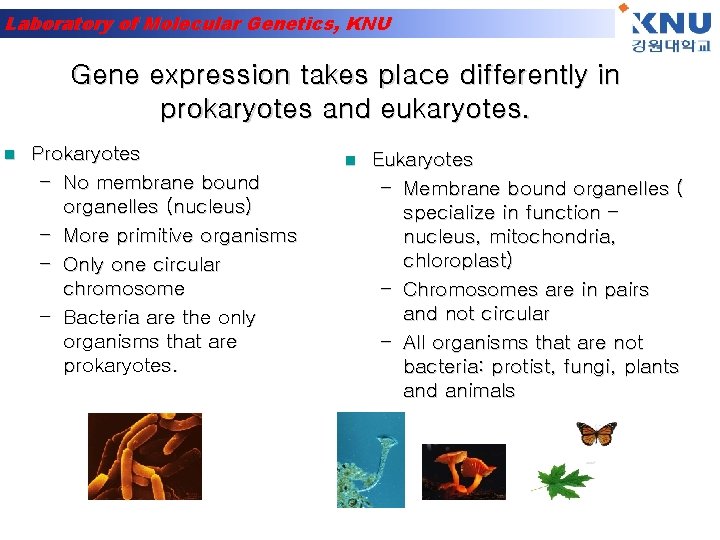
Laboratory of Molecular Genetics, KNU Gene expression takes place differently in prokaryotes and eukaryotes. n Prokaryotes – No membrane bound organelles (nucleus) – More primitive organisms – Only one circular chromosome – Bacteria are the only organisms that are prokaryotes. n Eukaryotes – Membrane bound organelles ( specialize in function – nucleus, mitochondria, chloroplast) – Chromosomes are in pairs and not circular – All organisms that are not bacteria: protist, fungi, plants and animals
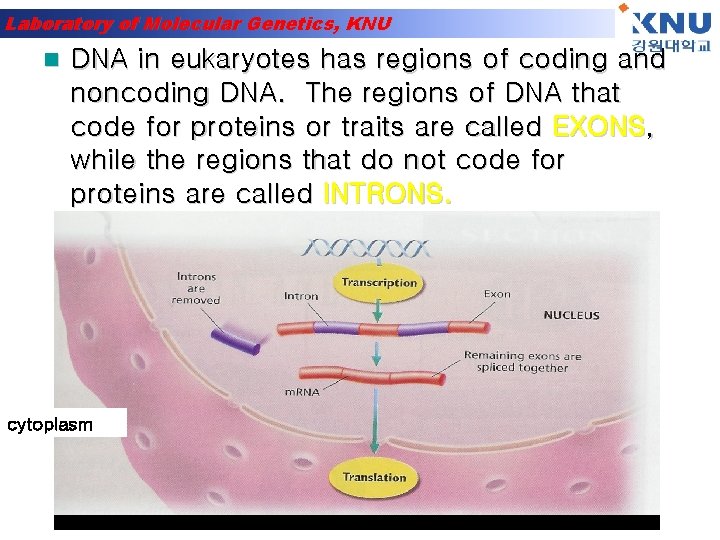
Laboratory of Molecular Genetics, KNU n DNA in eukaryotes has regions of coding and noncoding DNA. The regions of DNA that code for proteins or traits are called EXONS, while the regions that do not code for proteins are called INTRONS. cytoplasm
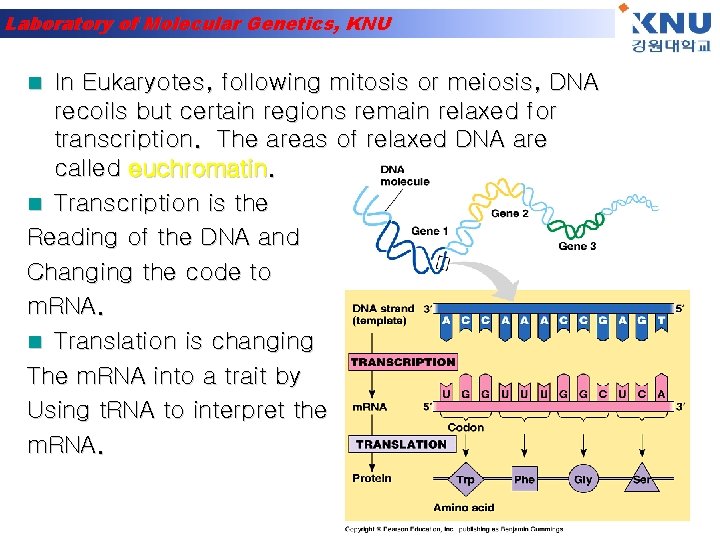
Laboratory of Molecular Genetics, KNU In Eukaryotes, following mitosis or meiosis, DNA recoils but certain regions remain relaxed for transcription. The areas of relaxed DNA are called euchromatin. n Transcription is the Reading of the DNA and Changing the code to m. RNA. n Translation is changing The m. RNA into a trait by Using t. RNA to interpret the m. RNA. n
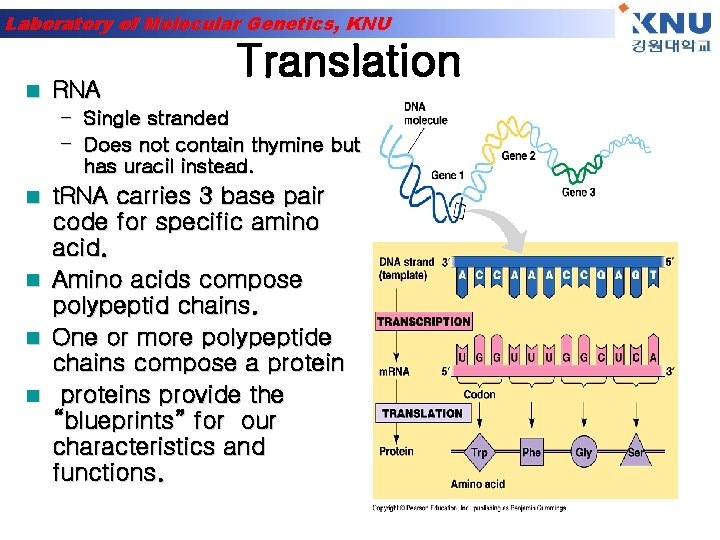
Laboratory of Molecular Genetics, KNU n RNA Translation – Single stranded – Does not contain thymine but has uracil instead. n n t. RNA carries 3 base pair code for specific amino acid. Amino acids compose polypeptid chains. One or more polypeptide chains compose a proteins provide the “blueprints” for our characteristics and functions.
Laboratory of Molecular Genetics, KNU
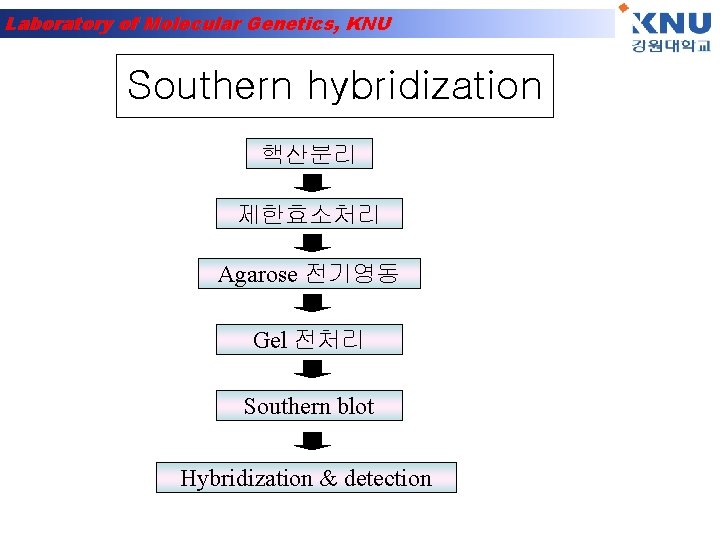
Laboratory of Molecular Genetics, KNU Southern hybridization 핵산분리 제한효소처리 Agarose 전기영동 Gel 전처리 Southern blot Hybridization & detection
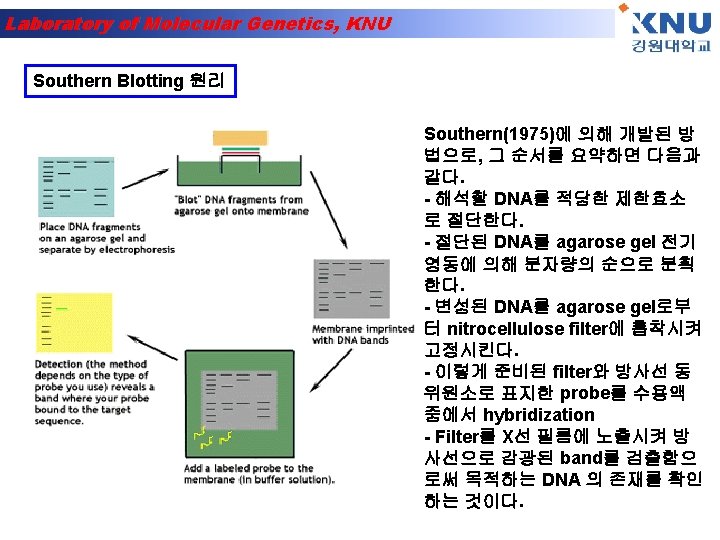

Laboratory of Molecular Genetics, KNU

Laboratory of Molecular Genetics, KNU

Laboratory of Molecular Genetics, KNU
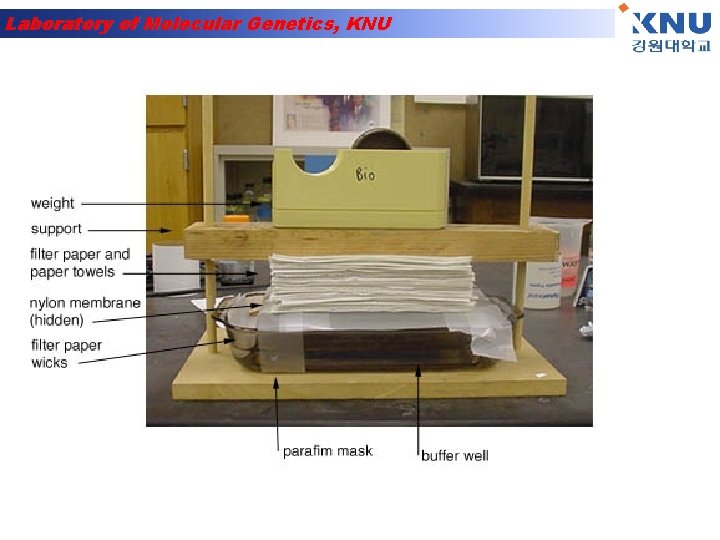
Laboratory of Molecular Genetics, KNU

Laboratory of Molecular Genetics, KNU
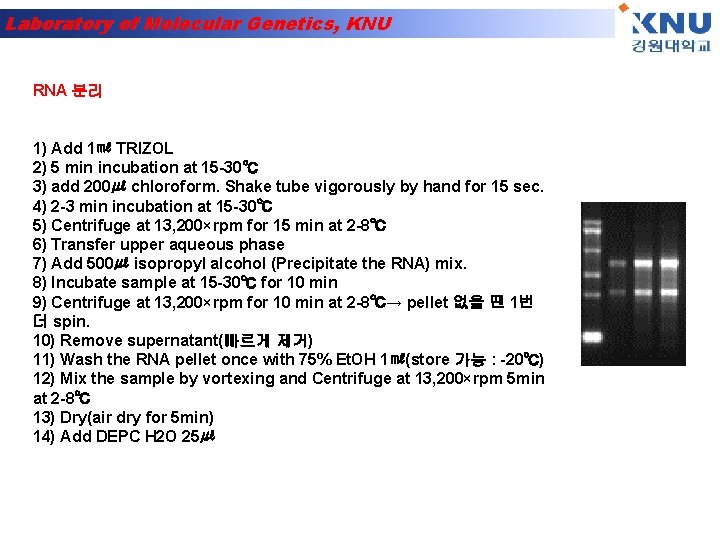
Laboratory of Molecular Genetics, KNU RNA 분리 1) Add 1㎖ TRIZOL 2) 5 min incubation at 15 -30℃ 3) add 200㎕ chloroform. Shake tube vigorously by hand for 15 sec. 4) 2 -3 min incubation at 15 -30℃ 5) Centrifuge at 13, 200×rpm for 15 min at 2 -8℃ 6) Transfer upper aqueous phase 7) Add 500㎕ isopropyl alcohol (Precipitate the RNA) mix. 8) Incubate sample at 15 -30℃ for 10 min 9) Centrifuge at 13, 200×rpm for 10 min at 2 -8℃→ pellet 없을 땐 1번 더 spin. 10) Remove supernatant(빠르게 제거) 11) Wash the RNA pellet once with 75% Et. OH 1㎖(store 가능 : -20℃) 12) Mix the sample by vortexing and Centrifuge at 13, 200×rpm 5 min at 2 -8℃ 13) Dry(air dry for 5 min) 14) Add DEPC H 2 O 25㎕
Laboratory of Molecular Genetics, KNU
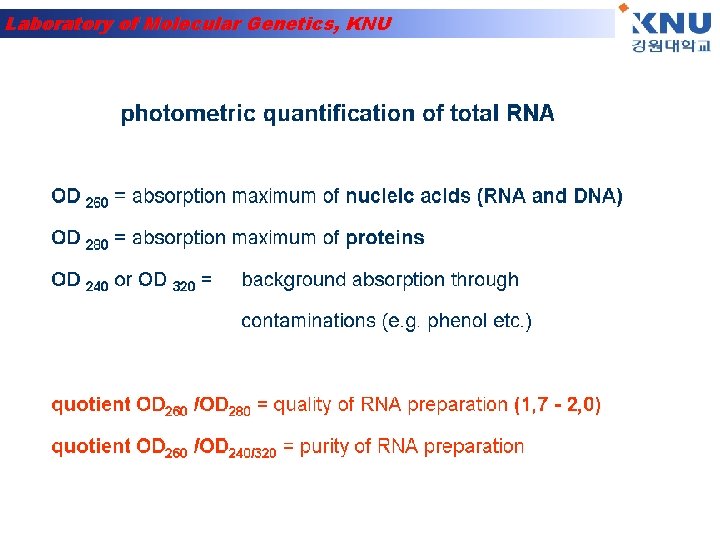
Laboratory of Molecular Genetics, KNU

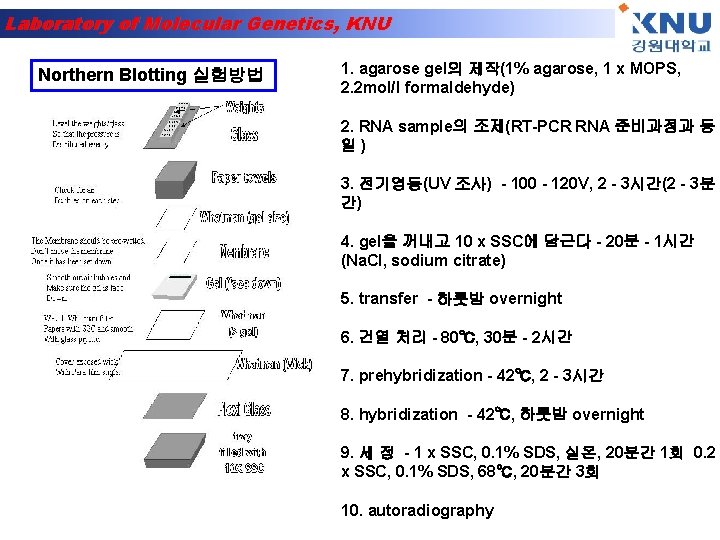
Laboratory of Molecular Genetics, KNU Northern Blotting 실험방법 1. agarose gel의 제작(1% agarose, 1 x MOPS, 2. 2 mol/l formaldehyde) 2. RNA sample의 조제(RT-PCR RNA 준비과정과 동 일) 3. 전기영동(UV 조사) - 100 - 120 V, 2 - 3시간(2 - 3분 간) 4. gel을 꺼내고 10 x SSC에 담근다 - 20분 - 1시간 (Na. Cl, sodium citrate) 5. transfer - 하룻밤 overnight 6. 건열 처리 - 80℃, 30분 - 2시간 7. prehybridization - 42℃, 2 - 3시간 8. hybridization - 42℃, 하룻밤 overnight 9. 세 정 - 1 x SSC, 0. 1% SDS, 실온, 20분간 1회 0. 2 x SSC, 0. 1% SDS, 68℃, 20분간 3회 10. autoradiography
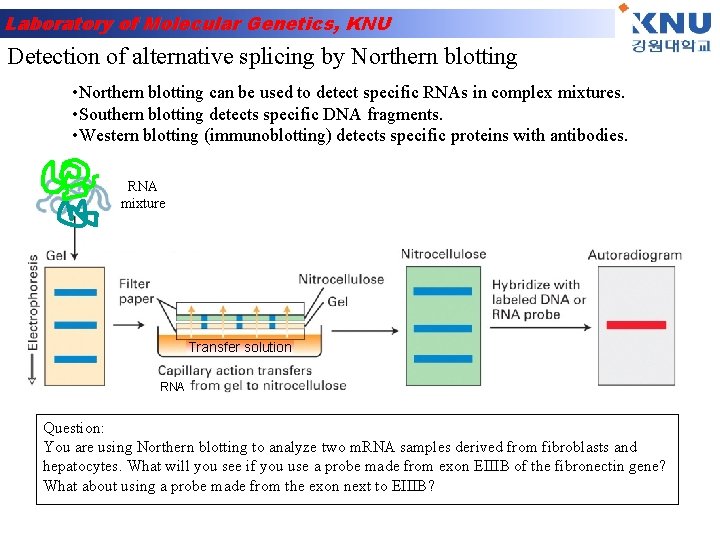
Laboratory of Molecular Genetics, KNU Detection of alternative splicing by Northern blotting • Northern blotting can be used to detect specific RNAs in complex mixtures. • Southern blotting detects specific DNA fragments. • Western blotting (immunoblotting) detects specific proteins with antibodies. RNA mixture Transfer solution RNA Question: You are using Northern blotting to analyze two m. RNA samples derived from fibroblasts and hepatocytes. What will you see if you use a probe made from exon EIIIB of the fibronectin gene? What about using a probe made from the exon next to EIIIB?


Laboratory of Molecular Genetics, KNU
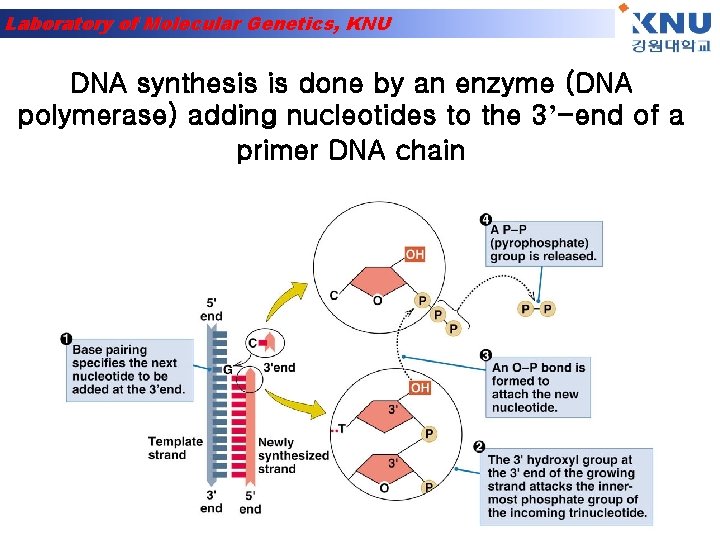
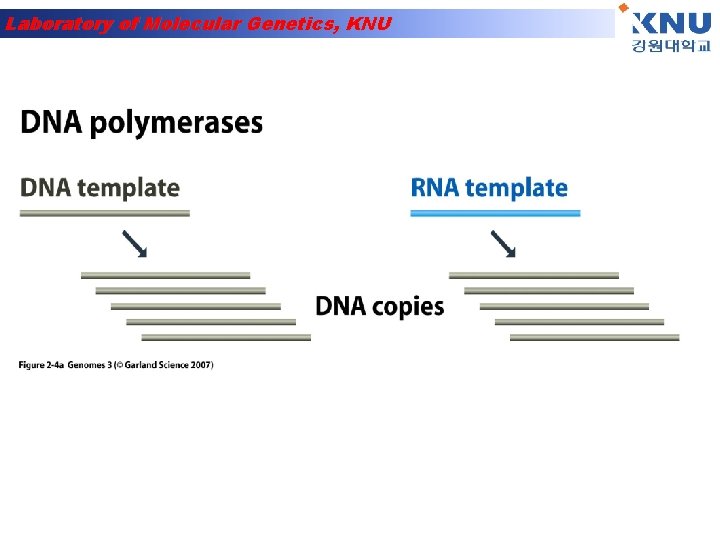
Laboratory of Molecular Genetics, KNU DNA synthesis is done by an enzyme (DNA polymerase) adding nucleotides to the 3’-end of a primer DNA chain
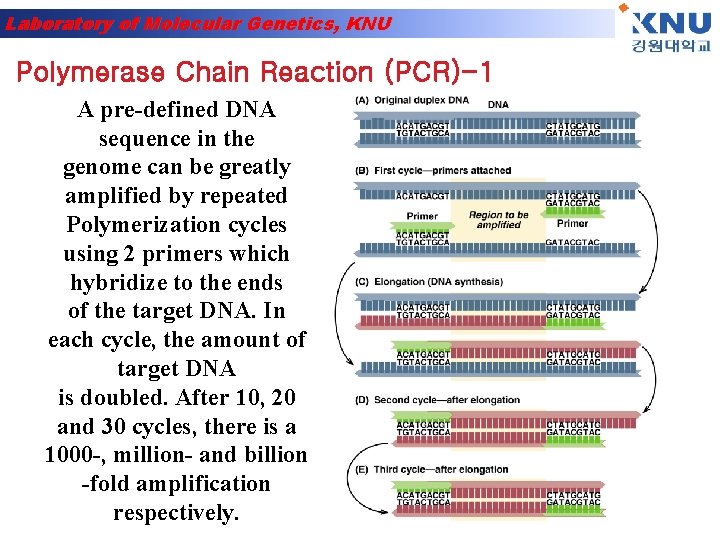
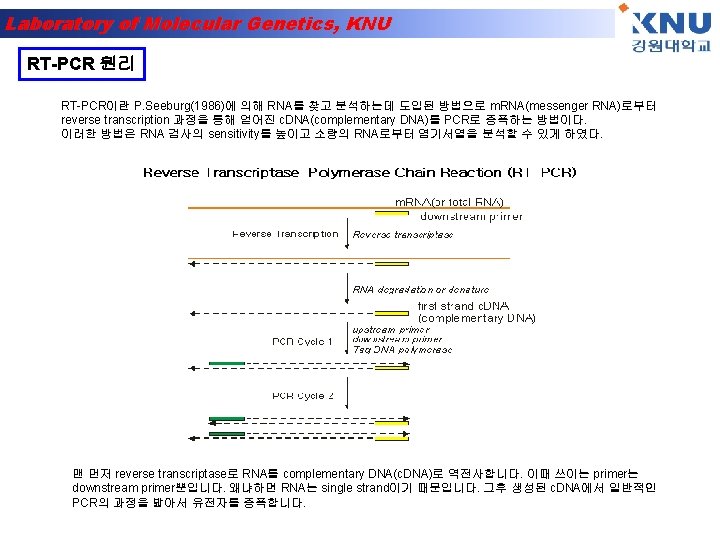
Laboratory of Molecular Genetics, KNU Polymerase Chain Reaction (PCR)-1 A pre-defined DNA sequence in the genome can be greatly amplified by repeated Polymerization cycles using 2 primers which hybridize to the ends of the target DNA. In each cycle, the amount of target DNA is doubled. After 10, 20 and 30 cycles, there is a 1000 -, million- and billion -fold amplification respectively.
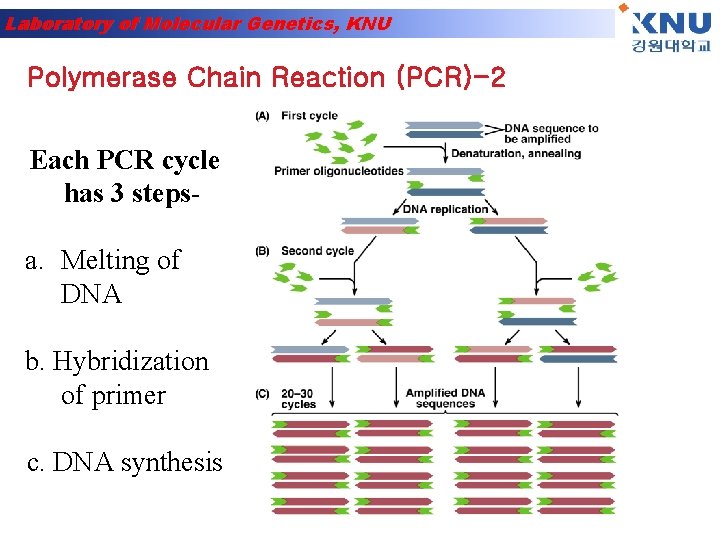
Laboratory of Molecular Genetics, KNU Polymerase Chain Reaction (PCR)-2 Each PCR cycle has 3 steps- a. Melting of DNA b. Hybridization of primer c. DNA synthesis
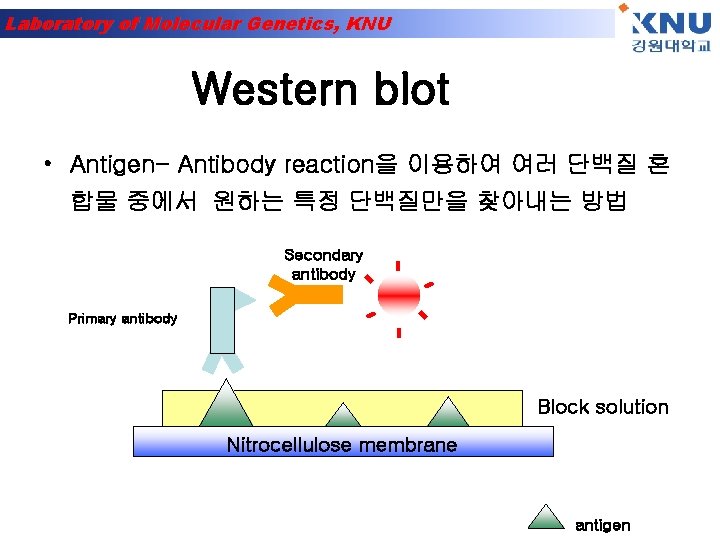
Laboratory of Molecular Genetics, KNU Western blot • Antigen- Antibody reaction을 이용하여 여러 단백질 혼 합물 중에서 원하는 특정 단백질만을 찾아내는 방법 Secondary antibody Primary antibody Block solution Nitrocellulose membrane antigen
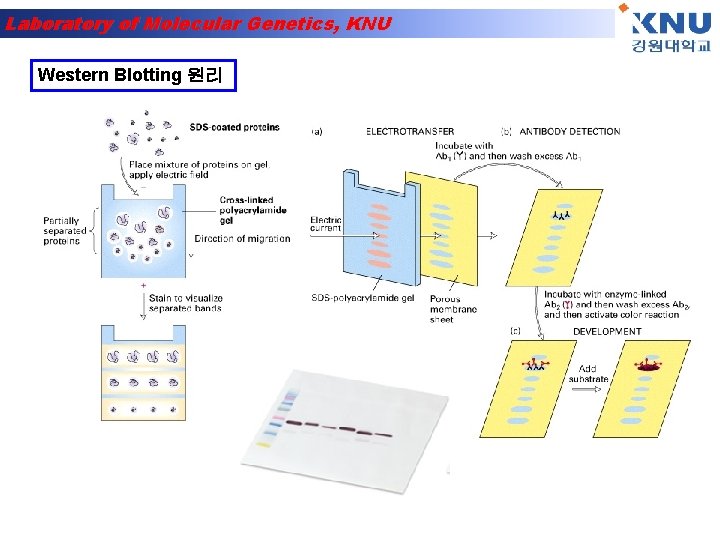
Laboratory of Molecular Genetics, KNU Western Blotting 원리
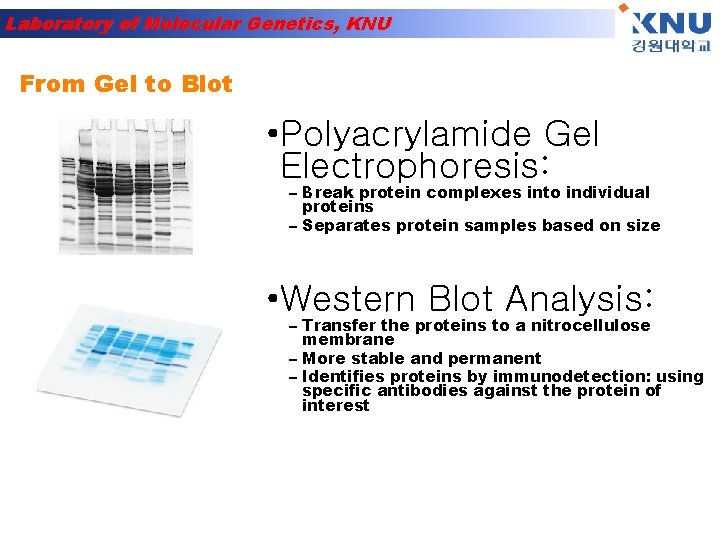
Laboratory of Molecular Genetics, KNU From Gel to Blot • Polyacrylamide Gel Electrophoresis: – Break protein complexes into individual proteins – Separates protein samples based on size • Western Blot Analysis: – Transfer the proteins to a nitrocellulose membrane – More stable and permanent – Identifies proteins by immunodetection: using specific antibodies against the protein of interest
Laboratory of Molecular Genetics, KNU Mini Trans-Blot Transfer Cell
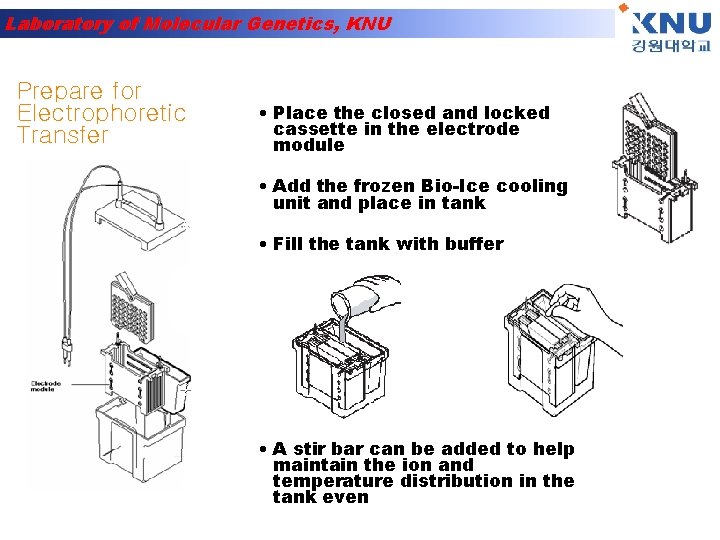
Laboratory of Molecular Genetics, KNU Prepare for Electrophoretic Transfer • Place the closed and locked cassette in the electrode module • Add the frozen Bio-Ice cooling unit and place in tank • Fill the tank with buffer • A stir bar can be added to help maintain the ion and temperature distribution in the tank even
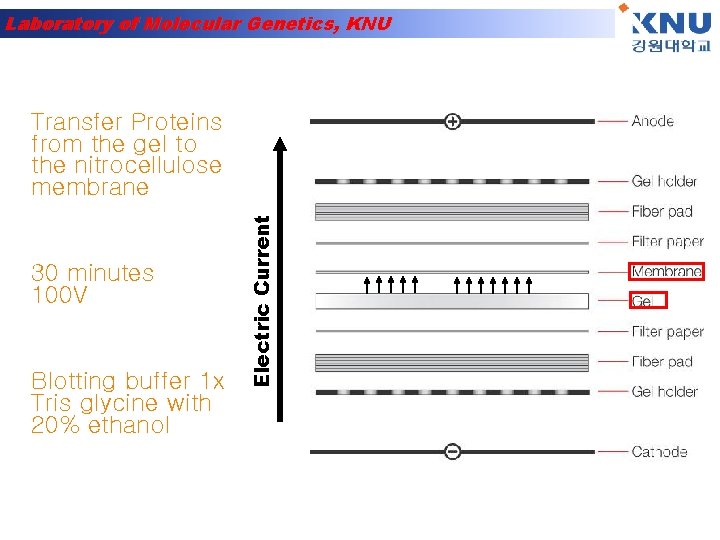
Laboratory of Molecular Genetics, KNU 30 minutes 100 V Blotting buffer 1 x Tris glycine with 20% ethanol Electric Current Transfer Proteins from the gel to the nitrocellulose membrane
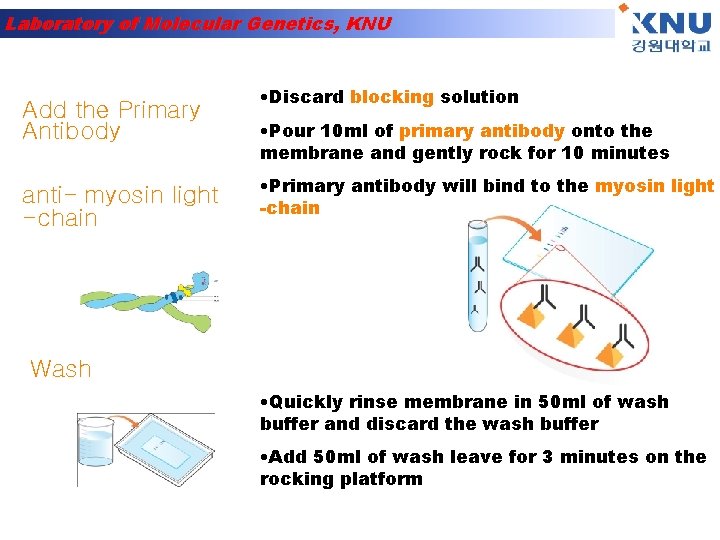
Laboratory of Molecular Genetics, KNU Add the Primary Antibody anti- myosin light -chain • Discard blocking solution • Pour 10 ml of primary antibody onto the membrane and gently rock for 10 minutes • Primary antibody will bind to the myosin light -chain Wash • Quickly rinse membrane in 50 ml of wash buffer and discard the wash buffer • Add 50 ml of wash leave for 3 minutes on the rocking platform
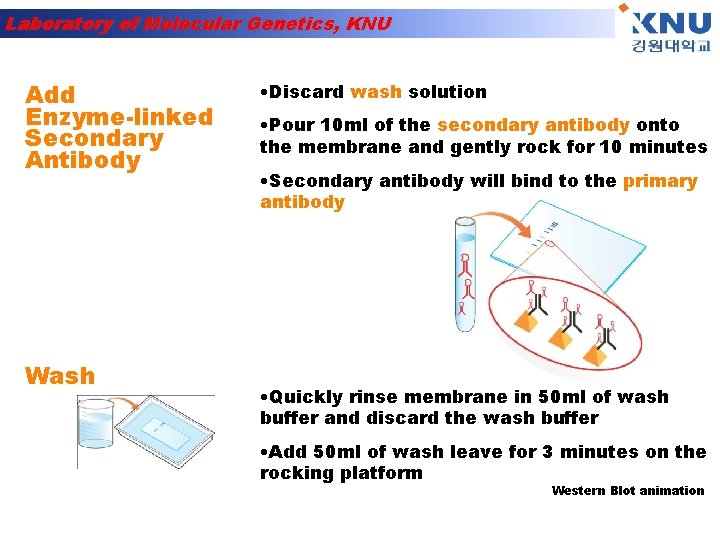
Laboratory of Molecular Genetics, KNU Add Enzyme-linked Secondary Antibody Wash • Discard wash solution • Pour 10 ml of the secondary antibody onto the membrane and gently rock for 10 minutes • Secondary antibody will bind to the primary antibody • Quickly rinse membrane in 50 ml of wash buffer and discard the wash buffer • Add 50 ml of wash leave for 3 minutes on the rocking platform Western Blot animation

Laboratory of Molecular Genetics, KNU Western Blotting 실험과정 A. SAMPLE PREPARATION Sample preparation procedures are provided for monolayer cells, suspension cells, and tissue samples. Follow the procedure suited to your needs. Monolayer Cells • Grow cells to subconfluency in a 100 mm x 20 mm petri dish, remove culture medium and rinse cell monolayer with room temperature 1 x PBS. The following steps should be performed on ice or at 4° C using fresh, ice cold buffers. • Add 0. 6 ml of RIPA buffer to the monolayer cells in the plate. Gently rock plate for 15 minutes at 4° C. Remove adherent cells with a cell scraper. Transfer the resulting lysate to a microcentrifuge tube. • Wash the plate once with 0. 3 ml of RIPA buffer and combine with first lysate. (Optional: Add 10 µl of 10 mg/ml PMSF and/or pass through a 21 -gauge needle to shear the DNA. ) Incubate 30– 60 minutes on ice. • Centrifuge cell lysate at 10, 000 xg for 10 minutes at 4° C. The supernatant fluid is the total cell lysate. Transfer the supernatant to a new microcentrifuge tube. This is your whole cell lysate. For increased protein recovery, resuspend the pellet in a small volume of RIPA, centrifuge and combine supernantants. Suspension Cells • Collect approximately 2. 0 x 107 cells by low-speed centrifugation (e. g. 200 xg) at room temperature for 5 minutes. Carefully remove culture medium. • Wash the pellet with PBS at room temperature, and again collect by low-speed centrifugation. Carefully remove supernatant. • Add 1. 0 ml of ice cold RIPA buffer with freshly added (Protease Inhibitors) and/or (Phosphatase Inhibitors). Gently resuspend cells in RIPA buffer with a pipet and incubate on ice for 30 minutes. • Further disrupt and homogenize cells by hydrodynamic shearing (21 -gauge needle), dounce homogenization or sonication, taking care not to raise the temperature of the lysate. (Optional: Add 10 µl of 10 mg/ml PMSF) Incubate 30 minutes on ice. • Transfer to microcentrifuge tube(s) and centrifuge at 10, 000 xg for 10 minutes at 4° C. The supernatant fluid is the total cell lysate. Transfer the supernatant to a new microfuge tube. This is your whole cell lysate. For increased protein recovery, resuspend the pellet in a small volume of RIPA, centrifuge and combine supernantants.

Laboratory of Molecular Genetics, KNU B. ELECTROPHORESIS • Mix sample (40– 60 µg whole cell lysate, 10– 20 µg nuclear extract or 10– 20 ng purified protein per lane) with an equal volume of 2 x electrophoresis sample buffer and boil for 2– 3 minutes. Unused samples may be stored at -20° C. • Load up to 10 µl of lysate per 1. 0 mm of well width for gels of 0. 75 mm thickness. • We recommend the use of Cruz Marker™ molecular weight standards. Load 2 µl/well for 0. 75 mm gels and 5 µl/well for 1. 5 mm gels. When used with Cruz Marker™ compatible secondary antibodies, internal standard bands will appear when the probed blot is exposed to detection reagent. Alternatively, use Prestained Molecular Weight Standards. • Electrophorese according to standard protocols. • Transfer proteins from the gel to a nitrocellulose or PVDF membrane using an electroblotting apparatus according to the manufacturer´s protocols. C. IMMUNOBLOTTING • Block non-specific binding by incubating membrane in Blotto (either Blotto A or Blotto B; Ig. G-free BSA, is recommended when using anti-bovine secondary antibodies) for 30– 60 minutes at room temperature. Alternatively, the membrane may be blocked at 4° C overnight in a covered container, using Blotto without Tween -20. • If using a phospho-specific antibody, add 0. 01% (v/v) of each Phosphatase Inhibitor Cocktails to the blocking solution and the antibody diluent to inhibit phosphatases. • Incubate the blocked membrane in primary antibody diluted in Blotto for 1 hour at room temperature. (For phospho-specific antibodies: Use Blotto B with 0. 01% (v/v) of each Phosphatase Inhibitor Cocktails Optimal antibody concentration should be determined by titration. We recommend a starting dilution of 0. 5 -2. 0 µg/ml. Wash membrane three times for 5 minutes each with TBST. • Incubate the membrane for 45 minutes at room temperature with horseradish peroxidase (HRP) conjugated secondary antibody, or alkaline phosphatase (AP) conjugated secondary antibody (Conventional Secondary Antibodies for Western Blotting), diluted to 1: 500– 1: 2000 in Blotto. If high backgrounds are observed, secondary antibody should be diluted further (up to 1: 20, 000). If Cruz Marker™ molecular weight standards are used in the gel, the Cruz Marker™ compatible secondary antibodies must be used in order to visualize standards with ECL. • Wash membrane three times for 5 minutes each with TBST and once for 5 minutes with TBS T. • Incubate membrane in Chemiluminescence Luminol Reagent according to Luminol data sheet, or visualize proteins

Laboratory of Molecular Genetics, KNU Transfection and Protein localization
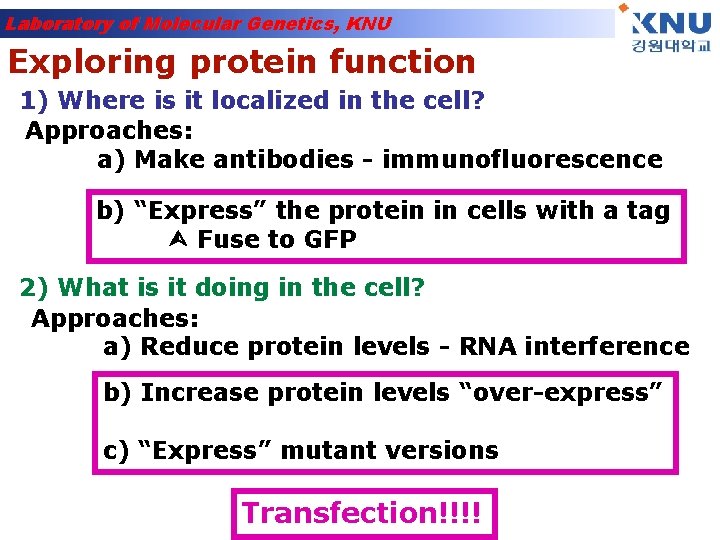
Laboratory of Molecular Genetics, KNU Exploring protein function 1) Where is it localized in the cell? Approaches: a) Make antibodies - immunofluorescence b) “Express” the protein in cells with a tag Fuse to GFP 2) What is it doing in the cell? Approaches: a) Reduce protein levels - RNA interference b) Increase protein levels “over-express” c) “Express” mutant versions Transfection!!!!

Laboratory of Molecular Genetics, KNU Transfection = Introduction of DNA into mammalian cells Gene is transcribed and translated into protein = “expressed”
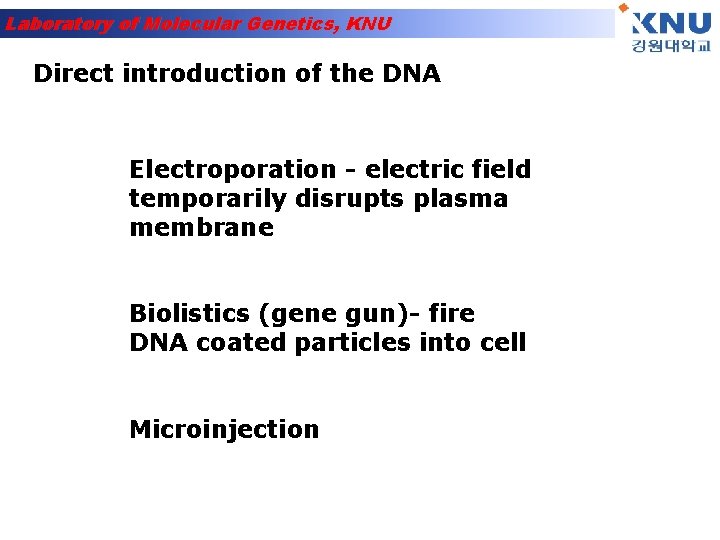
Laboratory of Molecular Genetics, KNU Direct introduction of the DNA Electroporation - electric field temporarily disrupts plasma membrane Biolistics (gene gun)- fire DNA coated particles into cell Microinjection

Laboratory of Molecular Genetics, KNU Virally-mediated introduction of the DNA Infection: Use recombinant viruses to deliver DNA Retroviruses Adenoviruses

Laboratory of Molecular Genetics, KNU Carrier-mediated introduction of the DNA Positively charged carrier molecules are mixed with the DNA and added to cell culture media: Calcium Phosphate DEAE Dextran liposomes micelles Carrier-DNA complexes bind to plasma membrane and are taken up
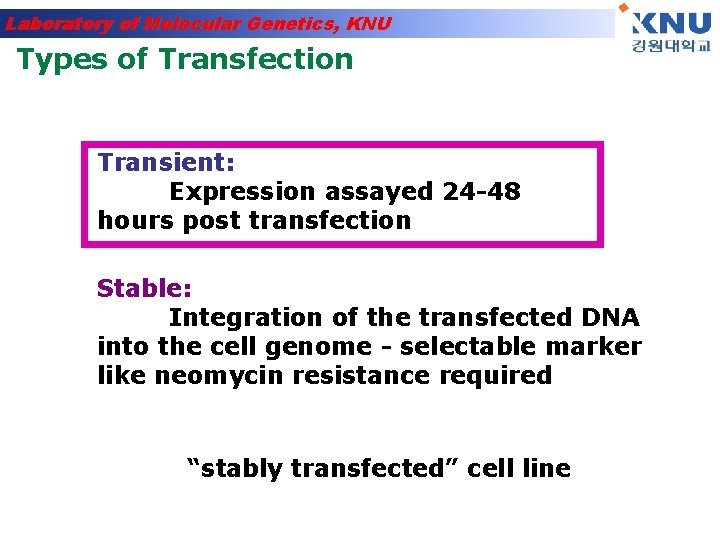
Laboratory of Molecular Genetics, KNU Types of Transfection Transient: Expression assayed 24 -48 hours post transfection Stable: Integration of the transfected DNA into the cell genome - selectable marker like neomycin resistance required “stably transfected” cell line
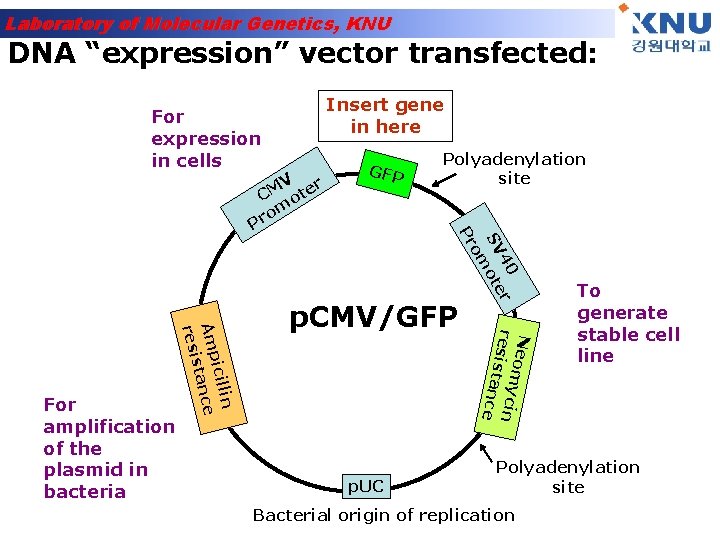
Laboratory of Molecular Genetics, KNU DNA “expression” vector transfected: Insert gene in here For expression in cells p. UC Ne o m ycin r e s i s tanc e p. CMV/GFP 40 r SV ote om n icilli Amp ance st resi For amplification of the plasmid in bacteria Polyadenylation site Pr V r CM ote om r P GFP To generate stable cell line Polyadenylation site Bacterial origin of replication
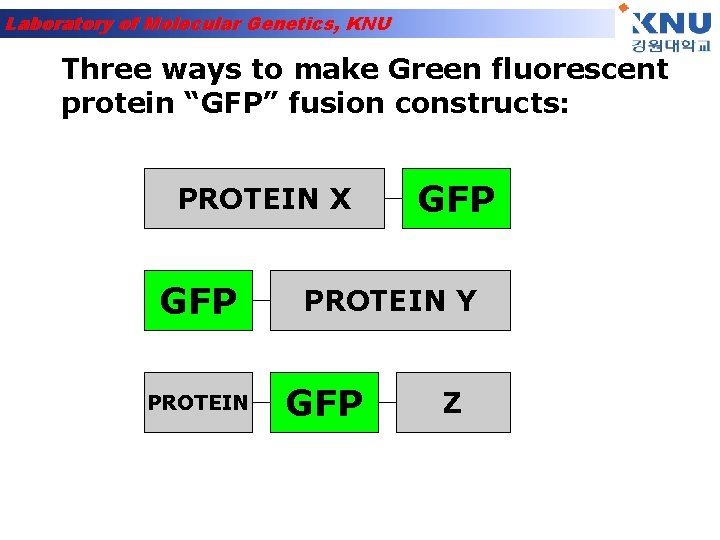
Laboratory of Molecular Genetics, KNU Three ways to make Green fluorescent protein “GFP” fusion constructs: PROTEIN X GFP PROTEIN Y GFP Z

Laboratory of Molecular Genetics, KNU EXPERIMENT: Transfect unknown GFP fusion protein Protein X, Y or Z Visualize GFP protein fluorescence by fluorescence microscopy in living cells Counter-stain with known marker to compare localization patterns in living cells = “vital stain”
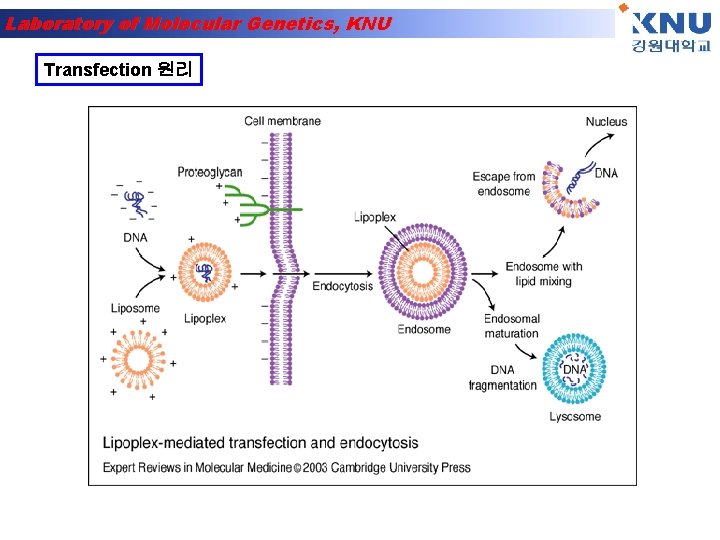
Laboratory of Molecular Genetics, KNU Transfection 원리
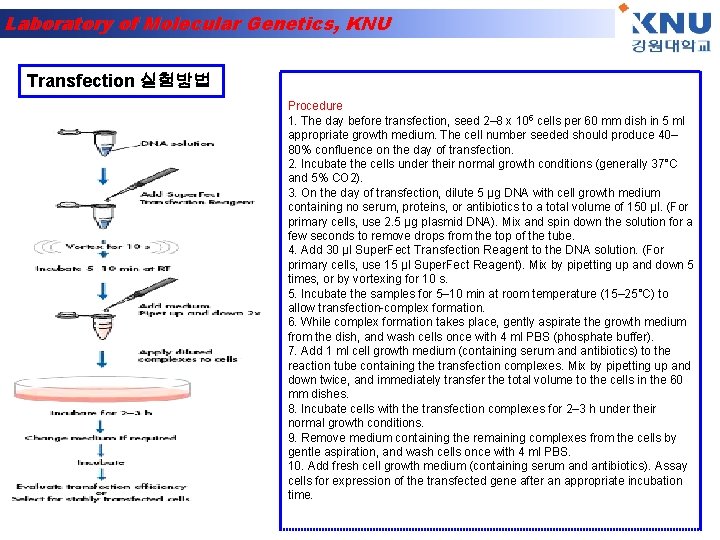
Laboratory of Molecular Genetics, KNU Transfection 실험방법 Procedure 1. The day before transfection, seed 2– 8 x 105 cells per 60 mm dish in 5 ml appropriate growth medium. The cell number seeded should produce 40– 80% confluence on the day of transfection. 2. Incubate the cells under their normal growth conditions (generally 37°C and 5% CO 2). 3. On the day of transfection, dilute 5 µg DNA with cell growth medium containing no serum, proteins, or antibiotics to a total volume of 150 µl. (For primary cells, use 2. 5 µg plasmid DNA). Mix and spin down the solution for a few seconds to remove drops from the top of the tube. 4. Add 30 µl Super. Fect Transfection Reagent to the DNA solution. (For primary cells, use 15 µl Super. Fect Reagent). Mix by pipetting up and down 5 times, or by vortexing for 10 s. 5. Incubate the samples for 5– 10 min at room temperature (15– 25°C) to allow transfection complex formation. 6. While complex formation takes place, gently aspirate the growth medium from the dish, and wash cells once with 4 ml PBS (phosphate buffer). 7. Add 1 ml cell growth medium (containing serum and antibiotics) to the reaction tube containing the transfection complexes. Mix by pipetting up and down twice, and immediately transfer the total volume to the cells in the 60 mm dishes. 8. Incubate cells with the transfection complexes for 2– 3 h under their normal growth conditions. 9. Remove medium containing the remaining complexes from the cells by gentle aspiration, and wash cells once with 4 ml PBS. 10. Add fresh cell growth medium (containing serum and antibiotics). Assay cells for expression of the transfected gene after an appropriate incubation time.
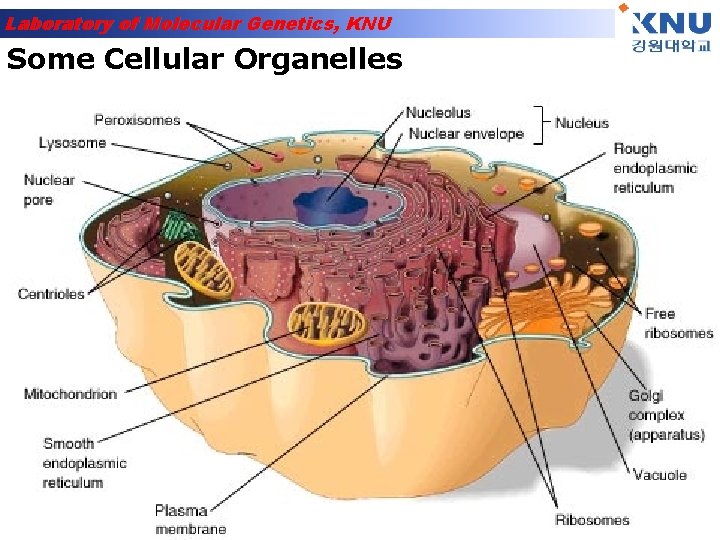
Laboratory of Molecular Genetics, KNU Some Cellular Organelles
Laboratory of Molecular Genetics, KNU • Compartments/organelles examined • Protein sequences sufficient for localization • Vital stains Nuclei Mitochondria Secretory Pathway: Endoplasmic Reticulum Golgi Complex Endocytotic Pathway: Endosomes
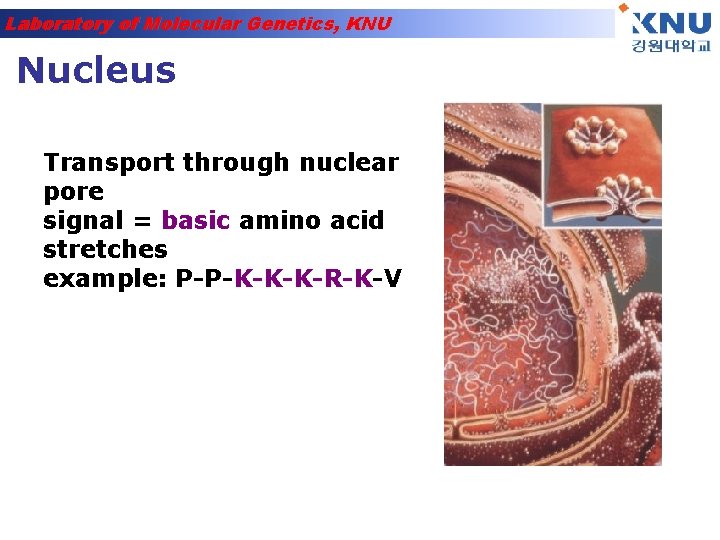
Laboratory of Molecular Genetics, KNU Nucleus Transport through nuclear pore signal = basic amino acid stretches example: P-P-K-K-K-R-K-V
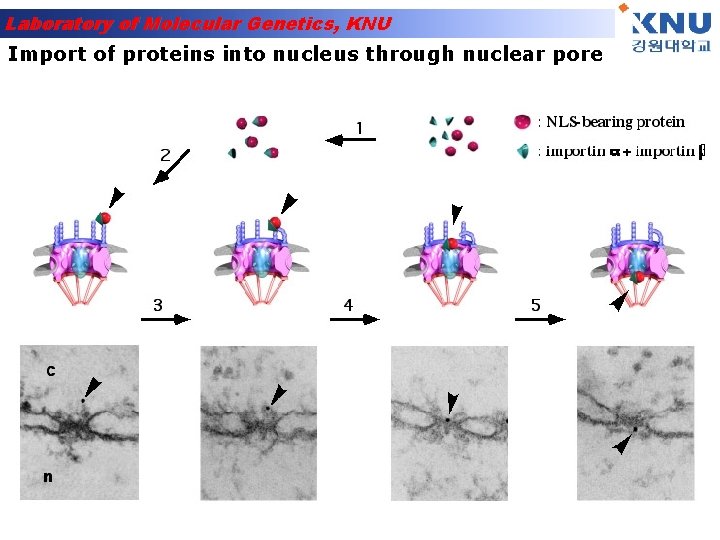
Laboratory of Molecular Genetics, KNU Import of proteins into nucleus through nuclear pore
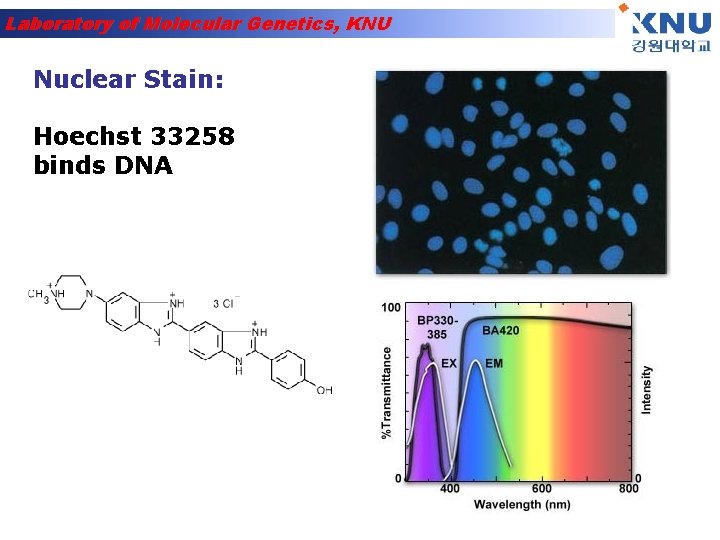
Laboratory of Molecular Genetics, KNU Nuclear Stain: Hoechst 33258 binds DNA
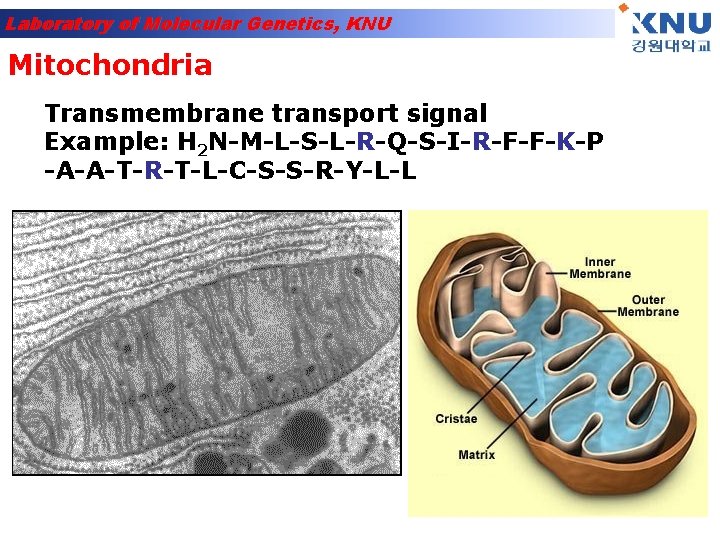
Laboratory of Molecular Genetics, KNU Mitochondria Transmembrane transport signal Example: H 2 N-M-L-S-L-R-Q-S-I-R-F-F-K-P -A-A-T-R-T-L-C-S-S-R-Y-L-L
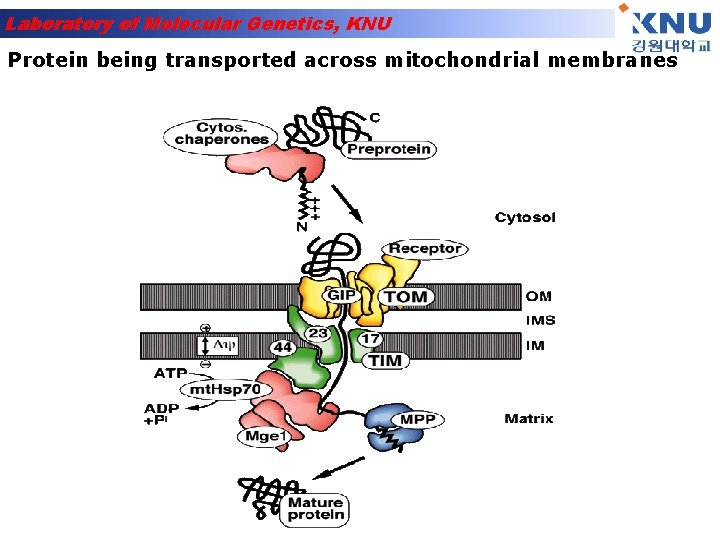
Laboratory of Molecular Genetics, KNU Protein being transported across mitochondrial membranes
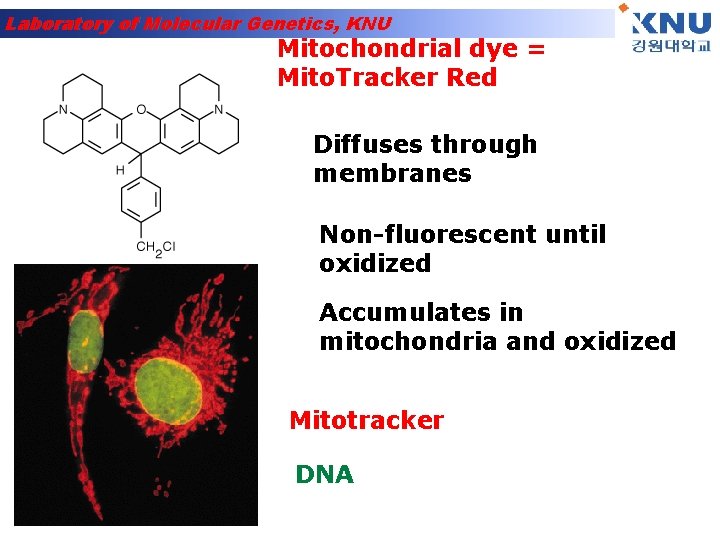
Laboratory of Molecular Genetics, KNU Mitochondrial dye = Mito. Tracker Red Diffuses through membranes Non-fluorescent until oxidized Accumulates in mitochondria and oxidized Mitotracker DNA
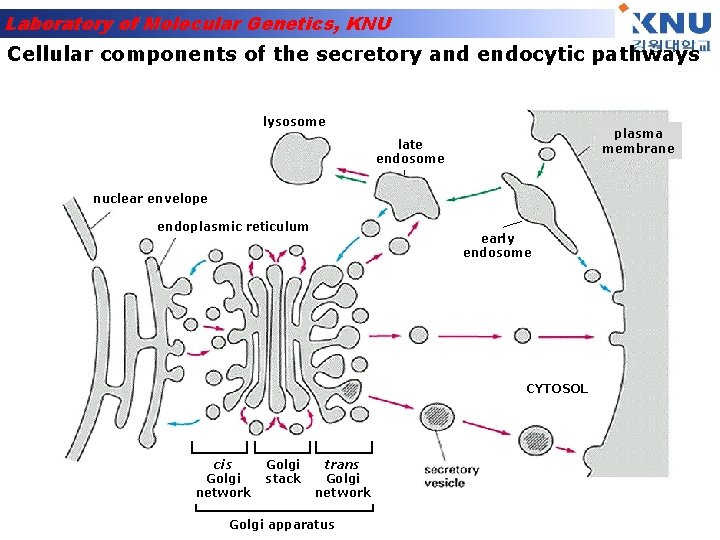
Laboratory of Molecular Genetics, KNU Cellular components of the secretory and endocytic pathways lysosome plasma membrane late endosome nuclear envelope endoplasmic reticulum early endosome CYTOSOL cis Golgi network Golgi stack trans Golgi network Golgi apparatus
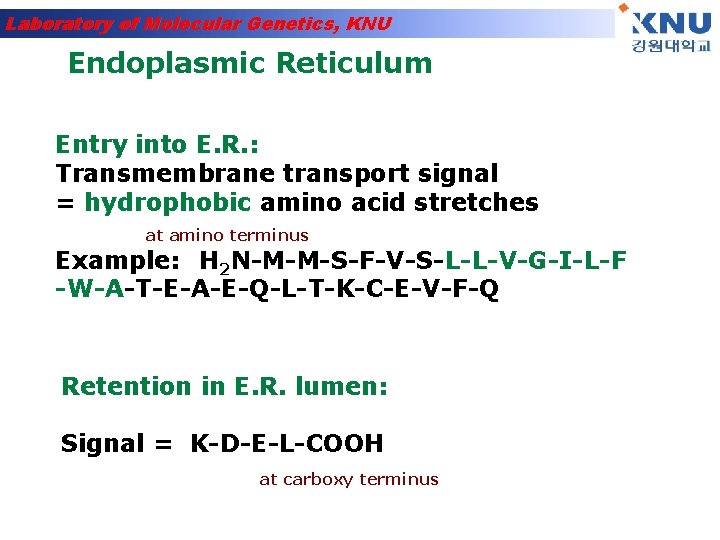
Laboratory of Molecular Genetics, KNU Endoplasmic Reticulum Entry into E. R. : Transmembrane transport signal = hydrophobic amino acid stretches at amino terminus Example: H 2 N-M-M-S-F-V-S-L-L-V-G-I-L-F -W-A-T-E-A-E-Q-L-T-K-C-E-V-F-Q Retention in E. R. lumen: Signal = K-D-E-L-COOH at carboxy terminus
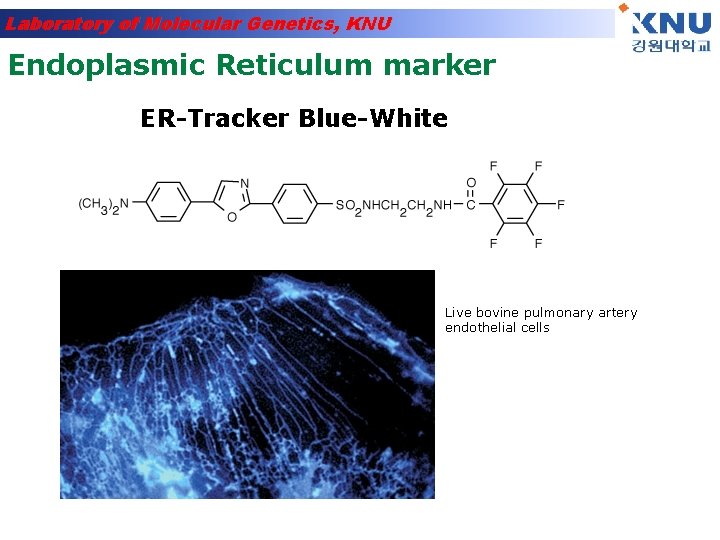
Laboratory of Molecular Genetics, KNU Endoplasmic Reticulum marker ER-Tracker Blue-White Live bovine pulmonary artery endothelial cells
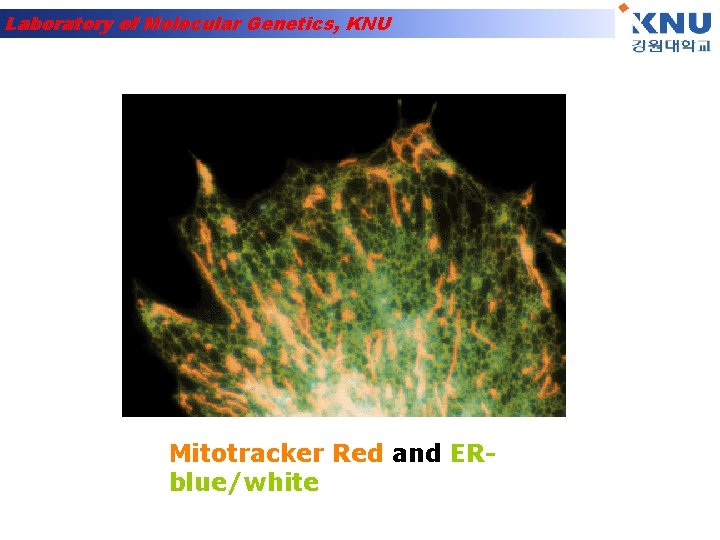
Laboratory of Molecular Genetics, KNU Mitotracker Red and ERblue/white
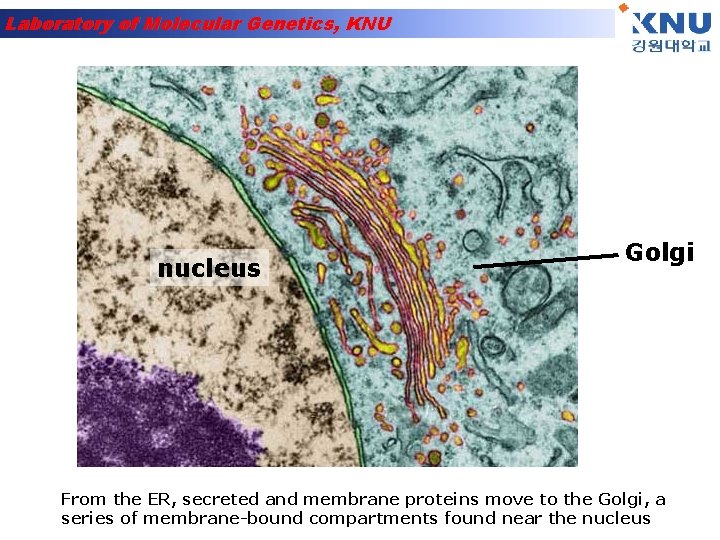
Laboratory of Molecular Genetics, KNU nucleus Golgi From the ER, secreted and membrane proteins move to the Golgi, a series of membrane-bound compartments found near the nucleus
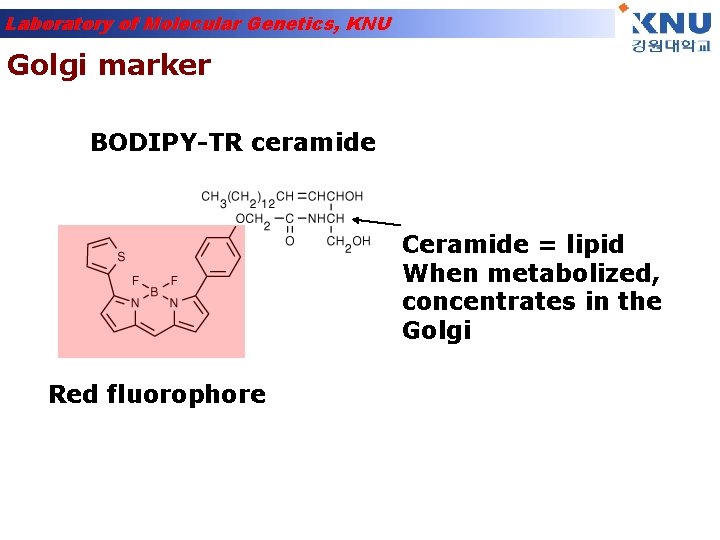
Laboratory of Molecular Genetics, KNU Golgi marker BODIPY-TR ceramide Ceramide = lipid When metabolized, concentrates in the Golgi Red fluorophore
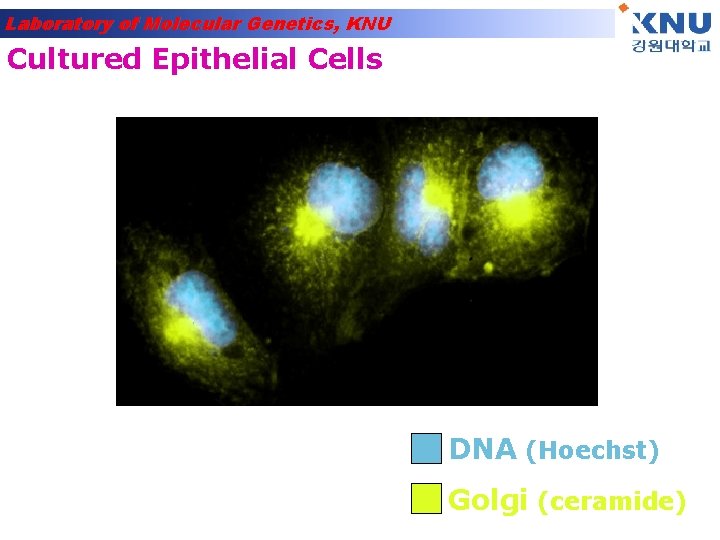
Laboratory of Molecular Genetics, KNU Cultured Epithelial Cells DNA (Hoechst) Golgi (ceramide)
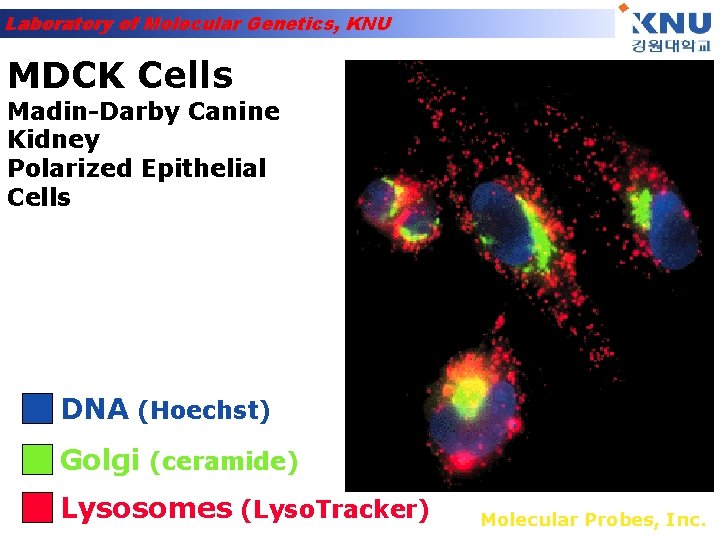
Laboratory of Molecular Genetics, KNU MDCK Cells Madin-Darby Canine Kidney Polarized Epithelial Cells DNA (Hoechst) Golgi (ceramide) Lysosomes (Lyso. Tracker) Molecular Probes, Inc.
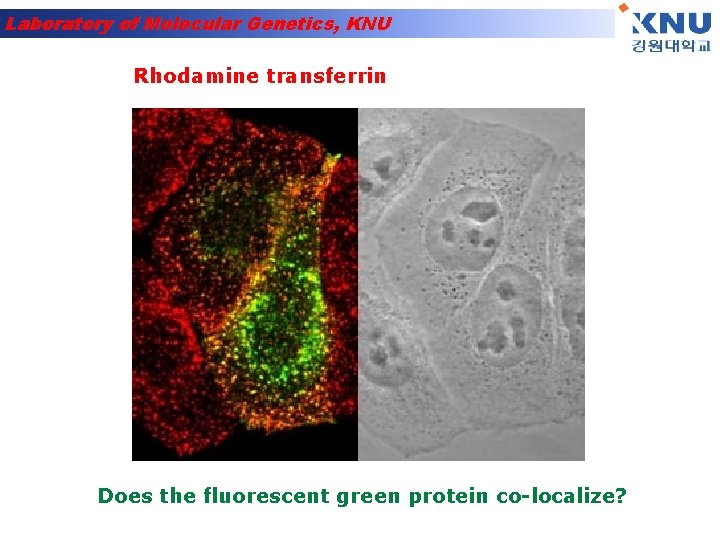
Laboratory of Molecular Genetics, KNU Rhodamine transferrin Does the fluorescent green protein co-localize?
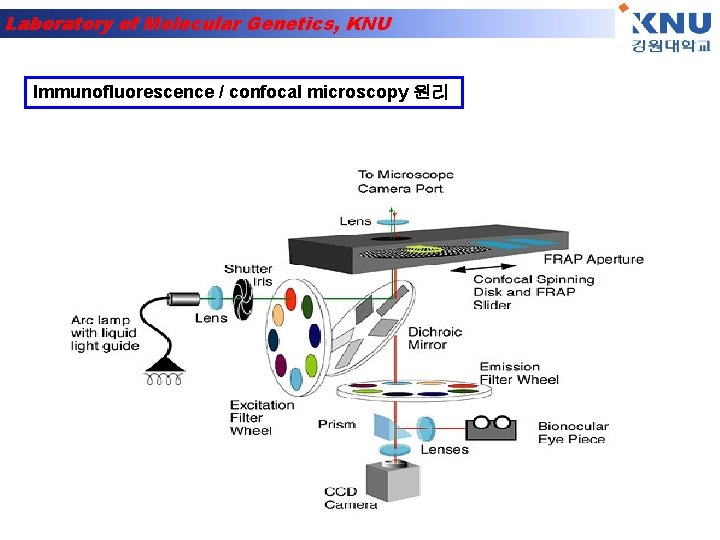
Laboratory of Molecular Genetics, KNU Immunofluorescence / confocal microscopy 원리

Laboratory of Molecular Genetics, KNU Immunofluorescence / confocal microscopy 실험방법 Immunofluorescence / confocal microscopy B or T cells in suspension, adherent cells on chambered coverglass or chamberslides, cryostat sections of unfixed, OCT embedded tissue: 1. wash cells 1 x cold RPMI (no wash for cryostat sections). 2. Fix 20 min 4% paraformaldehyde in 0. 1 M phosphate buffer p. H 7. 4, 0. 03 M sucrose on ice. ** 3. wash 2 x PBS/1%BSA. From now on everything can be at room temp. or on ice. 4. Permeabilize 5 min RT in 0. 2% saponin, PBS, 0. 03 M sucrose, 1% BSA. 5. wash 1 x PBS/BSA. 6. Block 15 min 5% normal goat serum (NGS) in PBS/BSA. 7. wash 1 x PBS/BSA. 8. 1° diluted in PBS/BSA 60 min RT; 100 l per tube or section. 9. wash 3 x PBS/BSA. 10. Block 15 min 5% NGS in PBS/BSA. 11. wash 1 x PBS/BSA. 12. 2° diluted in PBS/BSA 30 min RT; 100 l per tube or section. 13. wash 3 x PBS/BSA; (wash 1 x in PBS/BSA then 2 x 10 min in Molecular Probes Slow. Fade Light buffer if using Slow Fade Light S 7461 to coversip); pellet cells and put up in two drops of Molecular Probes Slowfade Light antifade medium. Pipet about 15 m l on slide and coverslip. For chambered coverglass just put two three drops in each chamber after wash.
- Slides: 72