Lab Week 10 and 11 Identification of an

Lab Week 10 and 11 Identification of an Unknown Bacteria of the Urinary Tract

Lab Goals • To distinguish between Escherichia coli, Proteus vulgaris, Pseudomonas aeruginosa, Staphylococcus epidermidis and Enterococcus faecalis on the basis of metabolic and exoenzyme testing • The tests used include: • • The Gram stain Mac. Conkey Agar Eosin-Methylene Blue Agar Indole Production Phenylalanine Deamination Nitrate Reduction Catalase

Lab Format • For each test this slide set provides • • Background Biochemistry Example Observations Expected Results (Controls) • To write your report (5% of your lab grade) • • Review the lab manual and this slide deck (click on videos and links) You will receive a set of observations to interpret into results Compare these results with those of the controls and complete the table Write your report according to the template provided

Filling Out the Table • Your observations (what you would see) will be provided • For example: Gram Stain Observation: Purple, round, clusters • Your results are the interpretation of those observations • Example: Result: Gram-positive staphylococci • The explanation of observations must include an explanation of the biochemical/enzymatic reaction that causes that result in your own words • Example Explanation: The bacteria have many layers of peptidoglycan in their cell wall that retain the crystal violet dye. • The organism/organisms indicated should include ALL species (in this case S. epidermidis, Enterococcus faecalis, Escherichia coli, Pseudomonas aeruginosa, and Proteus vulgaris) that would have that result. • Organisms Indicated: Staphylococcus epidermidis AND Enterococcus faecalis

The Gram Stain

Gram Positive and Gram Negative Cell Walls

Gram Staining • Primary Stain: Crystal Violet (+ charge) • Iodine: Mordant • Binds to dye, making it too large to pass through thick peptidoglycan • Alcohol: Decolorizer • Rinses away CV from Gram Negative Cells • Too much will cause the CV to leave even Gram Positive • Secondary Stain: Safranin (+ charge) • Remember, basic (+) dyes stain within the cell!
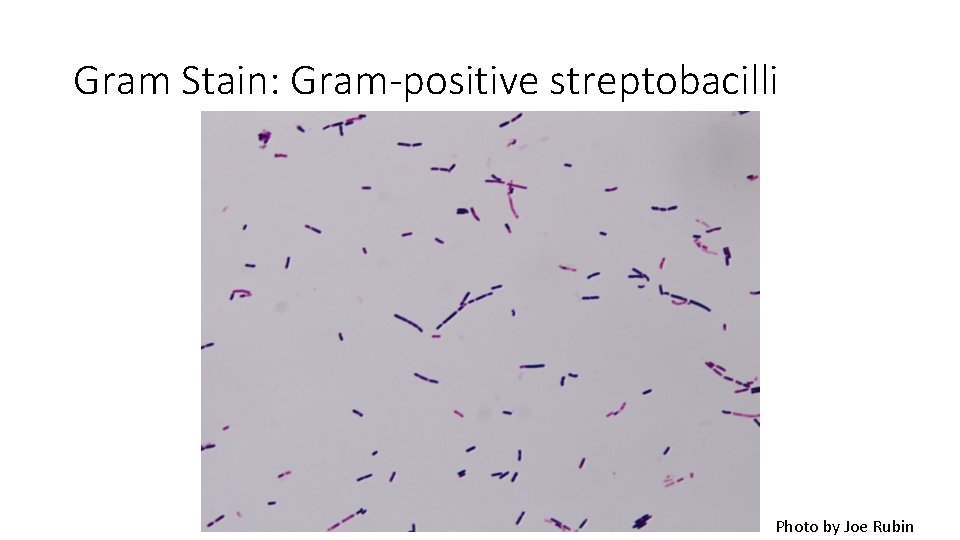
Gram Stain: Gram-positive streptobacilli Photo by Joe Rubin
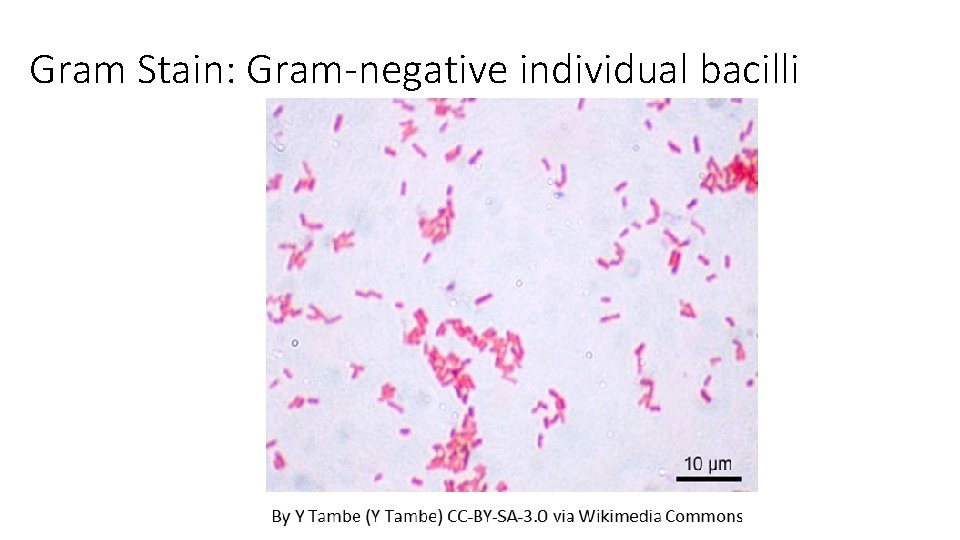
Gram Stain: Gram-negative individual bacilli
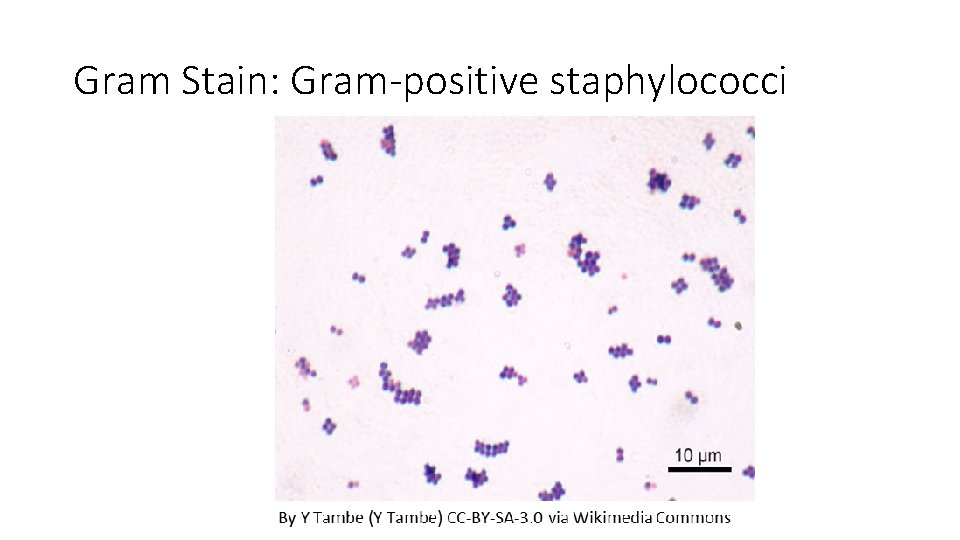
Gram Stain: Gram-positive staphylococci
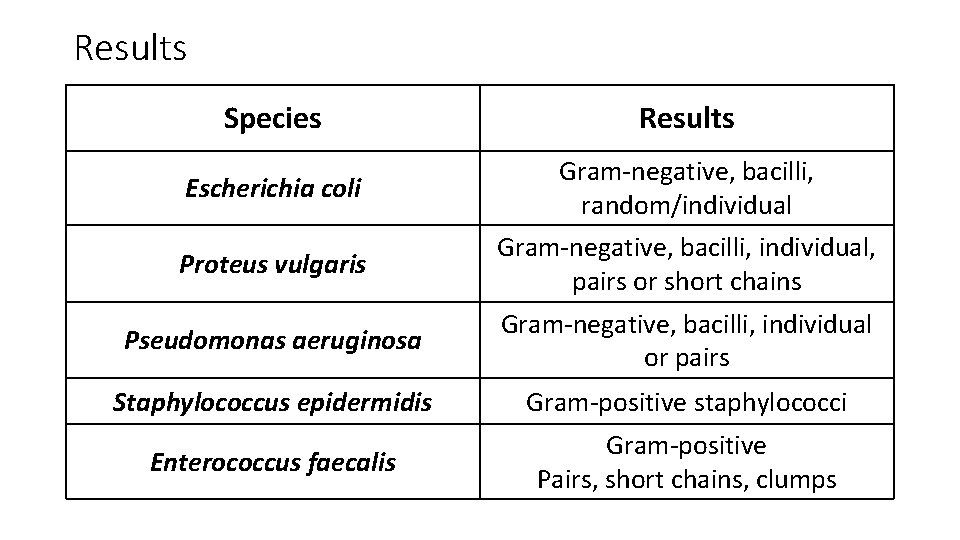
Results Species Results Escherichia coli Gram-negative, bacilli, random/individual Proteus vulgaris Gram-negative, bacilli, individual, pairs or short chains Pseudomonas aeruginosa Gram-negative, bacilli, individual or pairs Staphylococcus epidermidis Gram-positive staphylococci Enterococcus faecalis Gram-positive Pairs, short chains, clumps
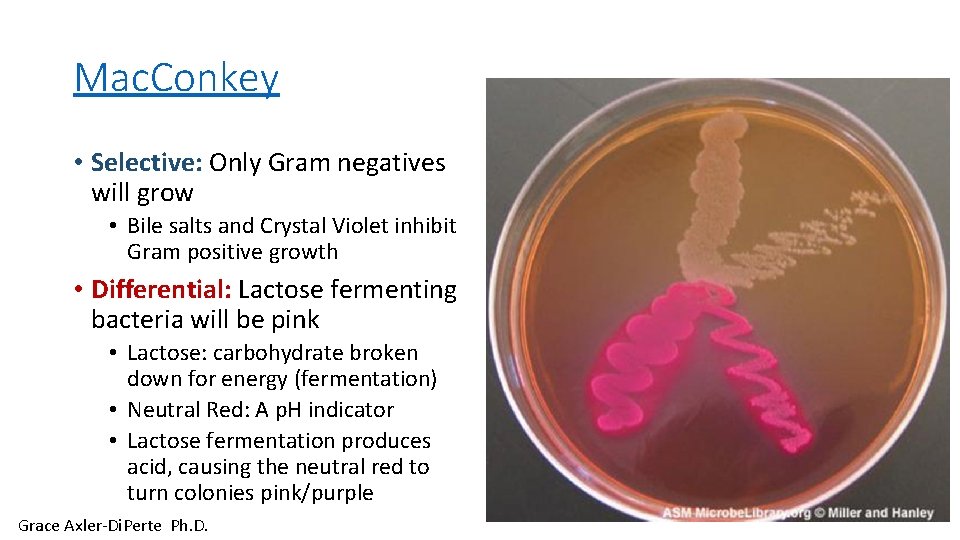
Mac. Conkey • Selective: Only Gram negatives will grow • Bile salts and Crystal Violet inhibit Gram positive growth • Differential: Lactose fermenting bacteria will be pink • Lactose: carbohydrate broken down for energy (fermentation) • Neutral Red: A p. H indicator • Lactose fermentation produces acid, causing the neutral red to turn colonies pink/purple Grace Axler-Di. Perte Ph. D.
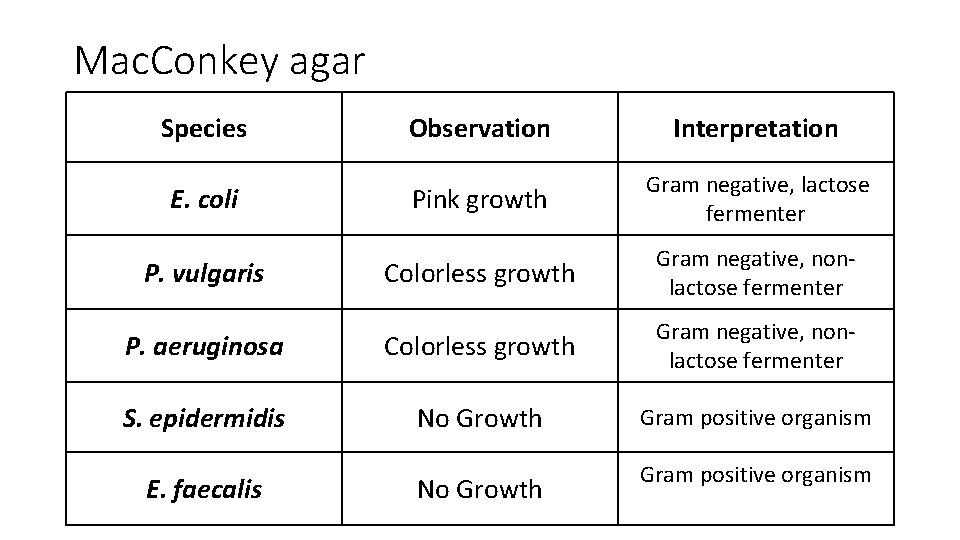
Mac. Conkey agar Species Observation Interpretation Pink growth Gram negative, lactose fermenter Colorless growth Gram negative, nonlactose fermenter P. aeruginosa Colorless growth Gram negative, nonlactose fermenter S. epidermidis No Growth Gram positive organism E. coli P. vulgaris E. faecalis

Eosin Methylene-Blue Agar • Selective: • Eosin and Methylene Blue are dyes which inhibit the growth of Gram positive organisms • Differential: • Lactose: carbohydrate broken down to make energy • Acid is produced lowering the p. H and causes the dues to precipitate • Lactose positive colonies will be metallic green Grace Axler-Di. Perte Ph. D. Joe Rubin (CC BY-NC-SA 2. 0)
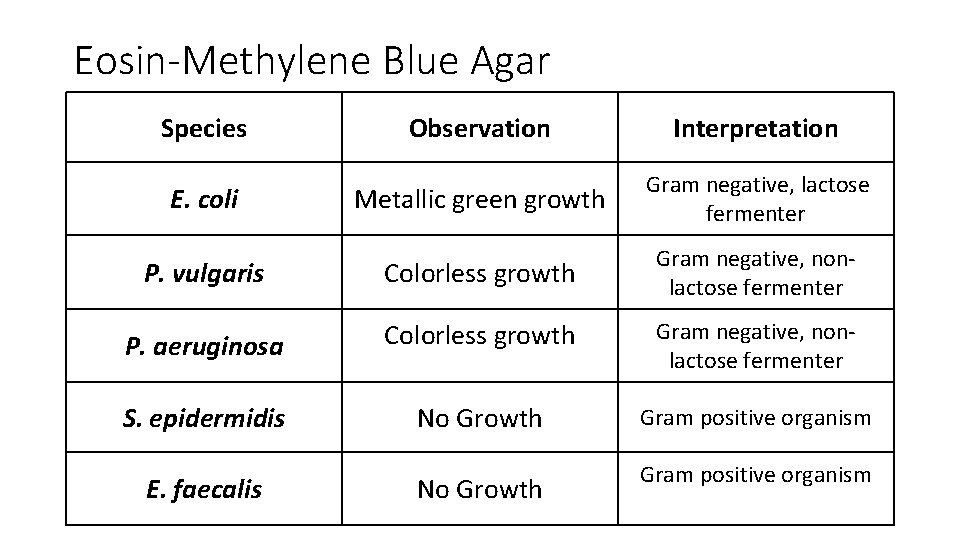
Eosin-Methylene Blue Agar Species Observation Interpretation Metallic green growth Gram negative, lactose fermenter P. vulgaris Colorless growth Gram negative, nonlactose fermenter P. aeruginosa Colorless growth Gram negative, nonlactose fermenter S. epidermidis No Growth Gram positive organism E. coli E. faecalis

Indole Production/Tryptophan Hydrolysis test

Tryptophan Hydrolysis (Indole Production) • Background and Biochemistry • Tryptophan Hydrolysis is a deamination reaction • Tryptophanase (enzyme) hydrolyzes tryptophan (aa) to produce indole (not used by the cell), pyruvic acid and ammonia (NH 3) (both used by the cell) • We detect indole using Kovac’s reagent, which reacts with indole to give a red compound • We use this test to distinguish E. coli from E. aerogenes Mary K. Ortiz, Ph. D.
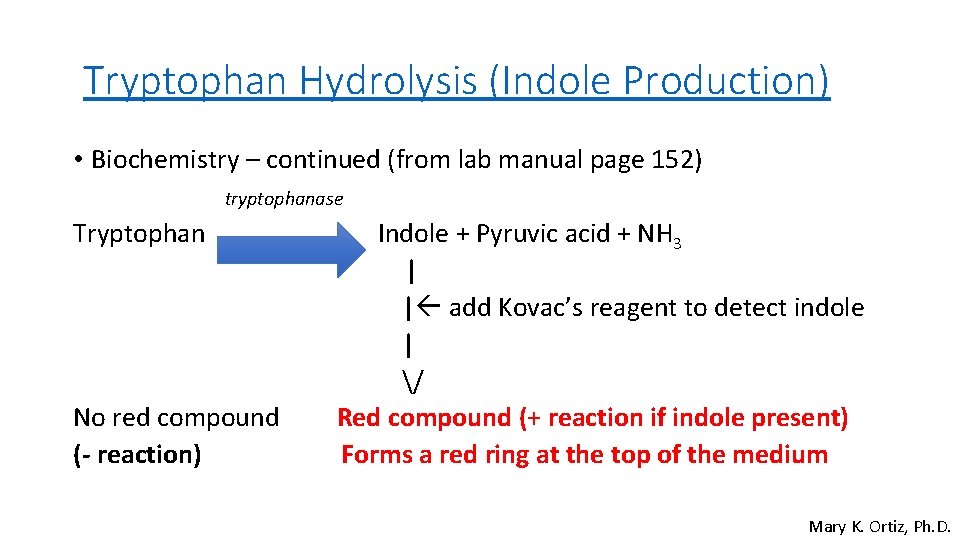
Tryptophan Hydrolysis (Indole Production) • Biochemistry – continued (from lab manual page 152) tryptophanase Tryptophan No red compound (- reaction) Indole + Pyruvic acid + NH 3 | | add Kovac’s reagent to detect indole | / Red compound (+ reaction if indole present) Forms a red ring at the top of the medium Mary K. Ortiz, Ph. D.

Tryptophan Hydrolysis (Indole Production) • Procedure (page 153 Lab Manual) • In this exercise five bacteria are used: • • • Escherichia coli (EC) Proteus vulgaris (PV) Pseudomonas aeruginosa (PA) Staphylococcus epidermidis (SE) Enterococcus faecalis (EF) • Each bacterium is inoculated into 1 tube of tryptone broth • • • Contains a high amount of tryptophan The tubes are incubated at 37 o. C (98. 6 o. F) for 24 -48 hours After incubation, add 10 drops of Kovac’s reagent and look for color change If tryptophanase present, then red ring forms at top of broth (+ result) If no red ring, no tryptophanase present (- result)

Tryptophan Hydrolysis (Indole Production) + -
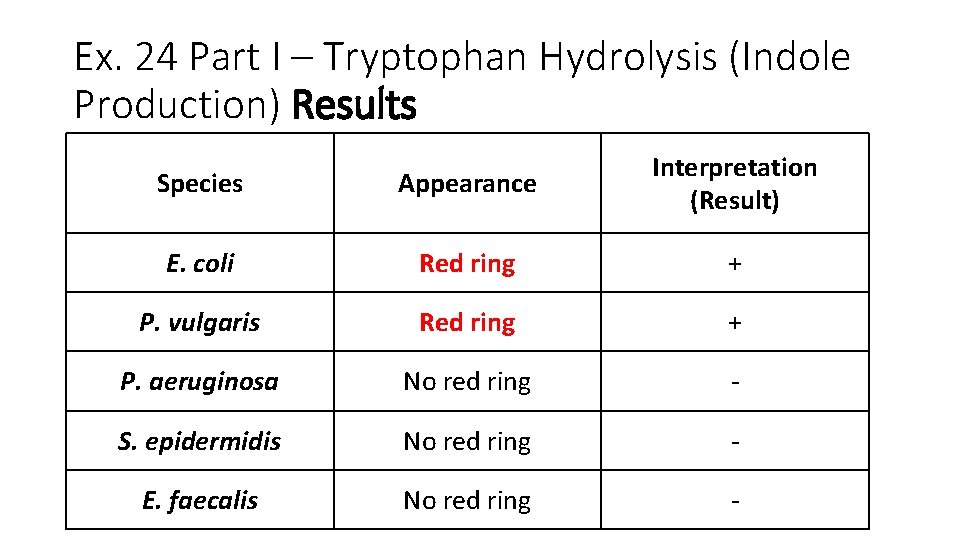
Ex. 24 Part I – Tryptophan Hydrolysis (Indole Production) Results Species Appearance Interpretation (Result) E. coli Red ring + P. vulgaris Red ring + P. aeruginosa No red ring - S. epidermidis No red ring - E. faecalis No red ring -

Ex. 24 Part II – Phenylalanine Deaminase • Background and Biochemistry • Phenylalanine (aa), with the enzyme phenylalanine deaminase (PAD) present gives phenylpyruvic acid and NH 3 • Some microbes use organic acids in reactions • We detect phenylpyruvic acid with ferric chloride (Fe. Cl 3) • Fe. Cl 3 reacts with phenylpyruvic acids to give a green color • Bacteria of the genus Proteus are all positive for PAD, so this is a useful test to test for this group of bacteria. Mary K. Ortiz, Ph. D.
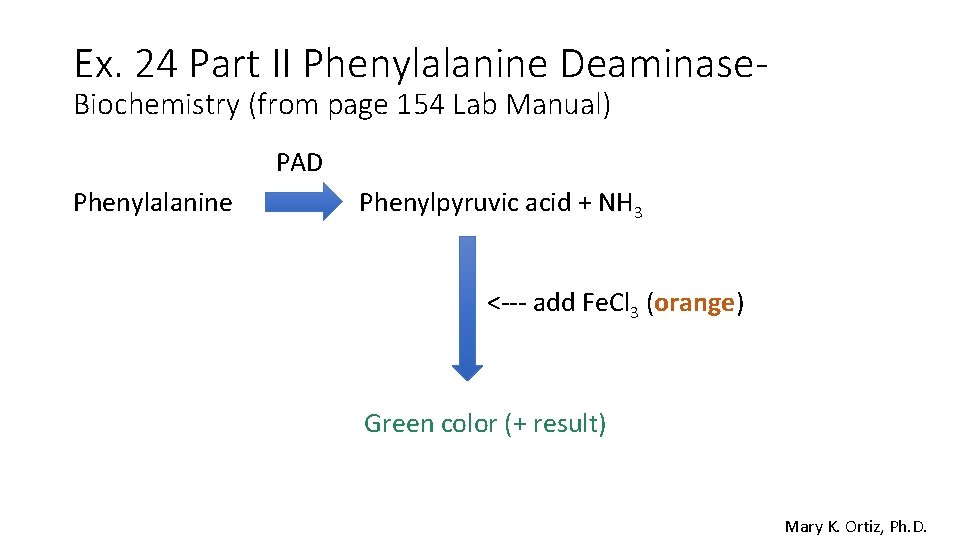
Ex. 24 Part II Phenylalanine Deaminase. Biochemistry (from page 154 Lab Manual) PAD Phenylalanine Phenylpyruvic acid + NH 3 <--- add Fe. Cl 3 (orange) Green color (+ result) Mary K. Ortiz, Ph. D.
Ex. 24 Part II Phenylalanine Deaminase • Procedure • In this exercise five bacteria are used: • • • E. coli (EC) Proteus vulgaris (PV) Pseudomonas aeruginosa (PA) Staphylococcus epidermidis (SE) Enterococcus faecalis (EF) • Each bacterium is inoculated into: • 1 Phenylalanine slant • • The tubes are incubated at 37 o. C (98. 6 o. F) for 24 -48 hours Add Fe. Cl 3 to the growth on the slant If PAD present, tube turns green (+ result) Otherwise, it is a – result (no color change)
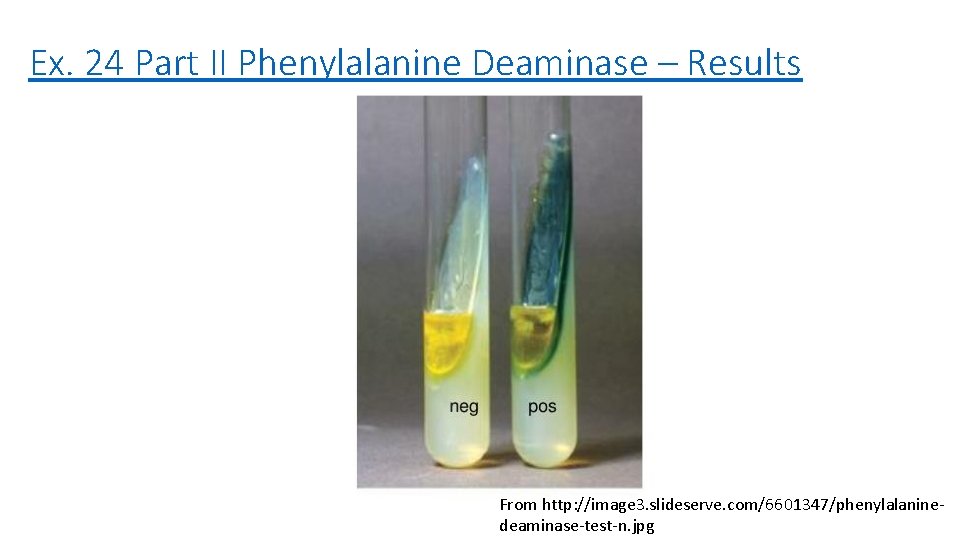
Ex. 24 Part II Phenylalanine Deaminase – Results From http: //image 3. slideserve. com/6601347/phenylalaninedeaminase-test-n. jpg
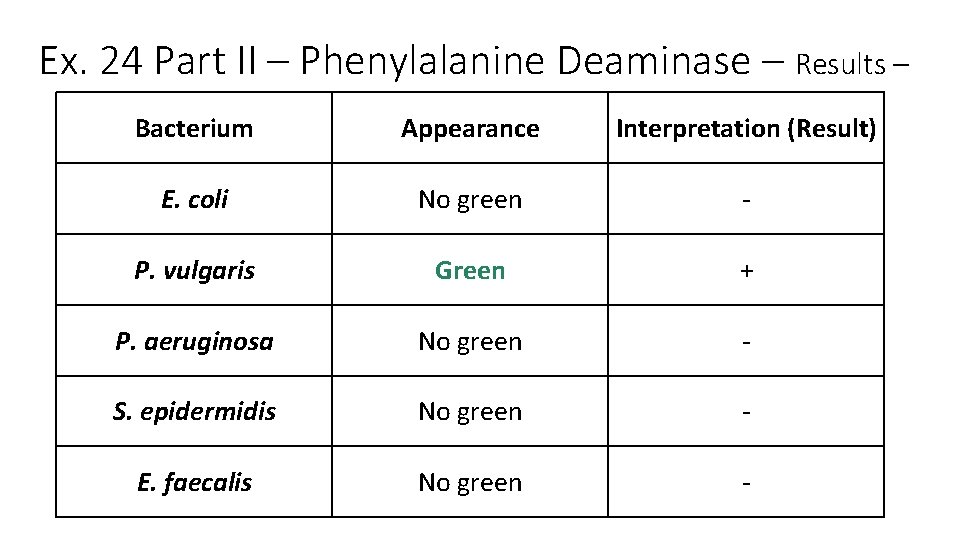
Ex. 24 Part II – Phenylalanine Deaminase – Results – Bacterium Appearance Interpretation (Result) E. coli No green - P. vulgaris Green + P. aeruginosa No green - S. epidermidis No green - E. faecalis No green -
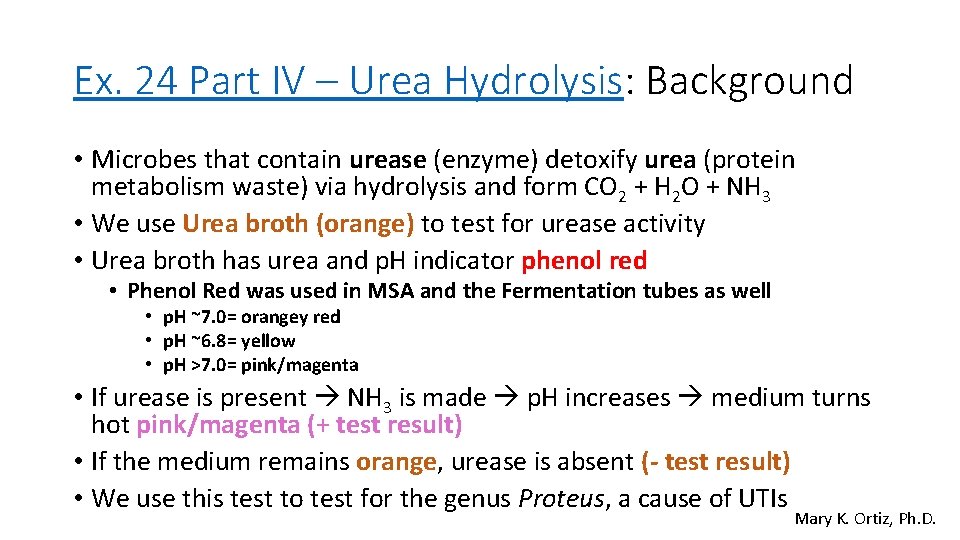
Ex. 24 Part IV – Urea Hydrolysis: Background • Microbes that contain urease (enzyme) detoxify urea (protein metabolism waste) via hydrolysis and form CO 2 + H 2 O + NH 3 • We use Urea broth (orange) to test for urease activity • Urea broth has urea and p. H indicator phenol red • Phenol Red was used in MSA and the Fermentation tubes as well • p. H ~7. 0= orangey red • p. H ~6. 8= yellow • p. H >7. 0= pink/magenta • If urease is present NH 3 is made p. H increases medium turns hot pink/magenta (+ test result) • If the medium remains orange, urease is absent (- test result) • We use this test to test for the genus Proteus, a cause of UTIs Mary K. Ortiz, Ph. D.
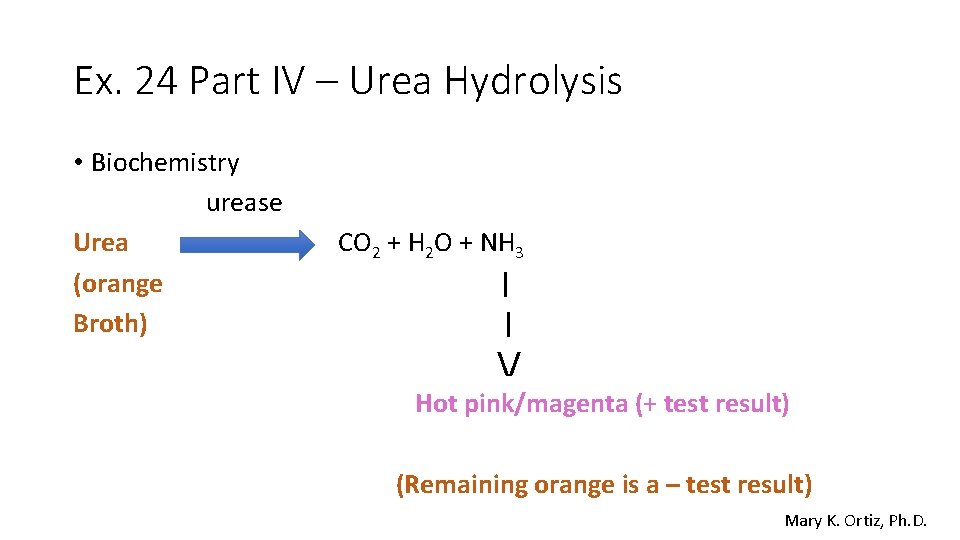
Ex. 24 Part IV – Urea Hydrolysis • Biochemistry urease Urea (orange Broth) CO 2 + H 2 O + NH 3 | | / Hot pink/magenta (+ test result) (Remaining orange is a – test result) Mary K. Ortiz, Ph. D.
Ex. 24 Part IV – Urea Hydrolysis: Procedure • In this exercise five bacteria are used: • • • E. coli (EC) Proteus vulgaris (PV) Pseudomonas aeruginosa (PA) Staphylococcus epidermidis (SE) Enterococcus faecalis (EF) • Each bacterium is inoculated into 1 tube of Urea broth • The tubes are incubated at 37 o. C (98. 6 o. F) for 5 days • In a positive test the broth turns hot pink/magenta • In a negative test, the broth remains orange
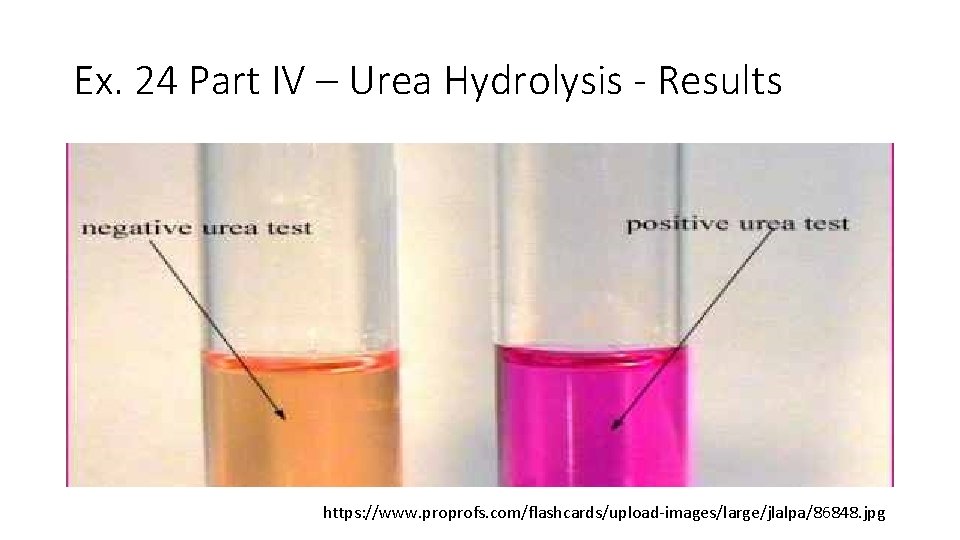
Ex. 24 Part IV – Urea Hydrolysis - Results https: //www. proprofs. com/flashcards/upload-images/large/jlalpa/86848. jpg
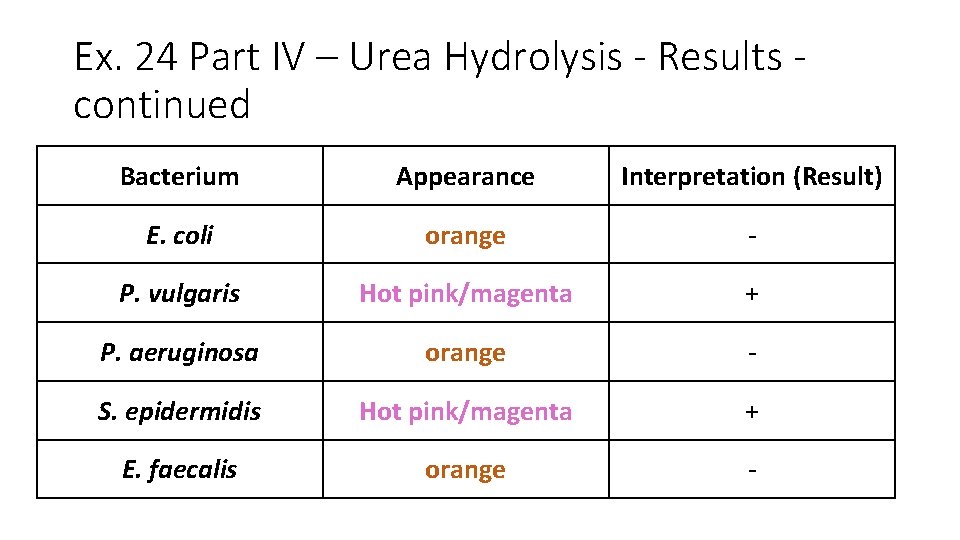
Ex. 24 Part IV – Urea Hydrolysis - Results continued Bacterium Appearance Interpretation (Result) E. coli orange - P. vulgaris Hot pink/magenta + P. aeruginosa orange - S. epidermidis Hot pink/magenta + E. faecalis orange -
Ex. 25 Part VI – Oxidase Activity • Background and Biochemistry • Cytochrome c oxidase has a main role in aerobic respirarion • It helps transport electrons from cytochrome c to O 2 (final eacceptor) • Oxidase reagent is used to test for oxidase activity • Oxidase reagent is poured onto the solid medium • If a microbe produces oxidase, the reagent turns from pink to black Mary K. Ortiz, Ph. D.
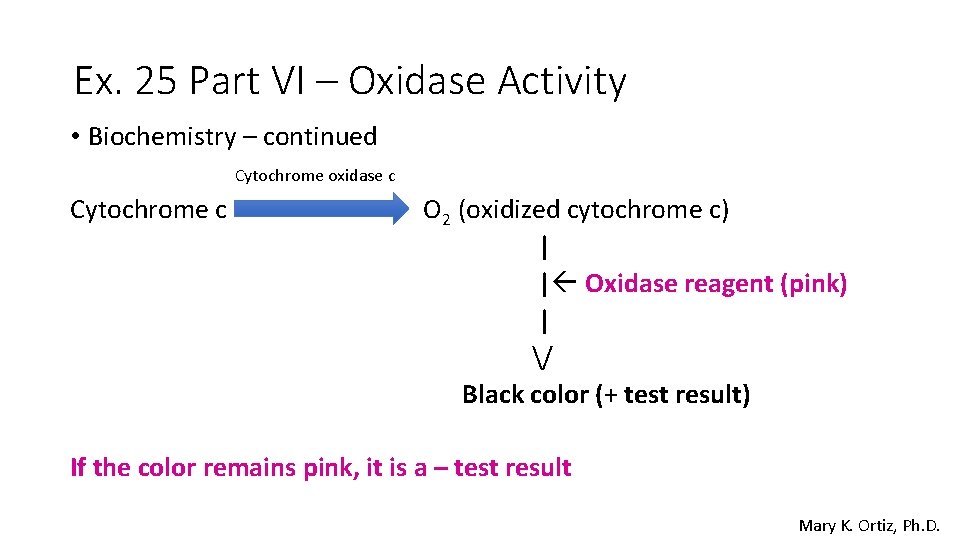
Ex. 25 Part VI – Oxidase Activity • Biochemistry – continued Cytochrome oxidase c Cytochrome c O 2 (oxidized cytochrome c) | | Oxidase reagent (pink) | / Black color (+ test result) If the color remains pink, it is a – test result Mary K. Ortiz, Ph. D.
Ex. 25 Part VI – Oxidase Activity: Procedure • In this exercise five bacteria are used: • • • E. coli (EC) Proteus vulgaris (PV) Pseudomonas aeruginosa (PA) Staphylococcus epidermidis (SE) Enterococcus faecalis (EF) • Each bacterium is inoculated onto a TSA (Tryptic Soy Agar) Plate • The tubes are incubated at 37 o. C (98. 6 o. F) for 24 -48 hours • Add the oxidase reagent to the growth on the plate or on a swab of the sample • If the plate turns blue-black, the microbe produces oxidase (+ test Result) • If the plate/swab remains pink, the microbe does not produce oxidase (test result)
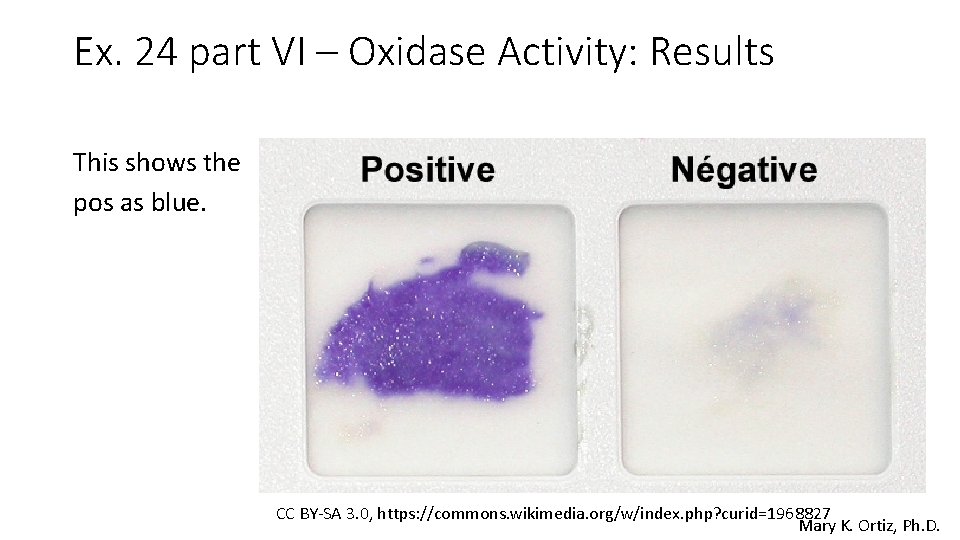
Ex. 24 part VI – Oxidase Activity: Results This shows the pos as blue. CC BY-SA 3. 0, https: //commons. wikimedia. org/w/index. php? curid=1968827 Mary K. Ortiz, Ph. D.
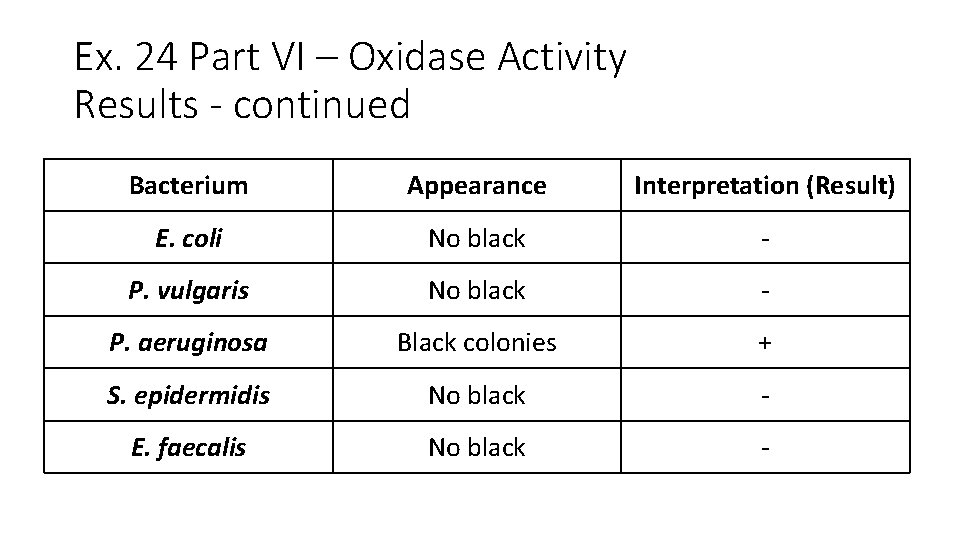
Ex. 24 Part VI – Oxidase Activity Results - continued Bacterium Appearance Interpretation (Result) E. coli No black - P. vulgaris No black - P. aeruginosa Black colonies + S. epidermidis No black - E. faecalis No black -
Ex. 25 Part VII – Nitrate Reduction: Background and Biochemistry • Some microbes use inorganic nitrate (NO 3) as a final e- acceptor • When NO 3 is used, it is reduced to nitrite (NO 2) by nitratase (enzyme) • Some microbes further reduce NO 2 to nitrogen gas (N 2) by nitritase (enzyme) in a reaction called denitrification • This video does not show total denitrification • Note the difference in spelling between nitratase and nitritase as they are two separate enzymes Mary K. Ortiz, Ph. D.
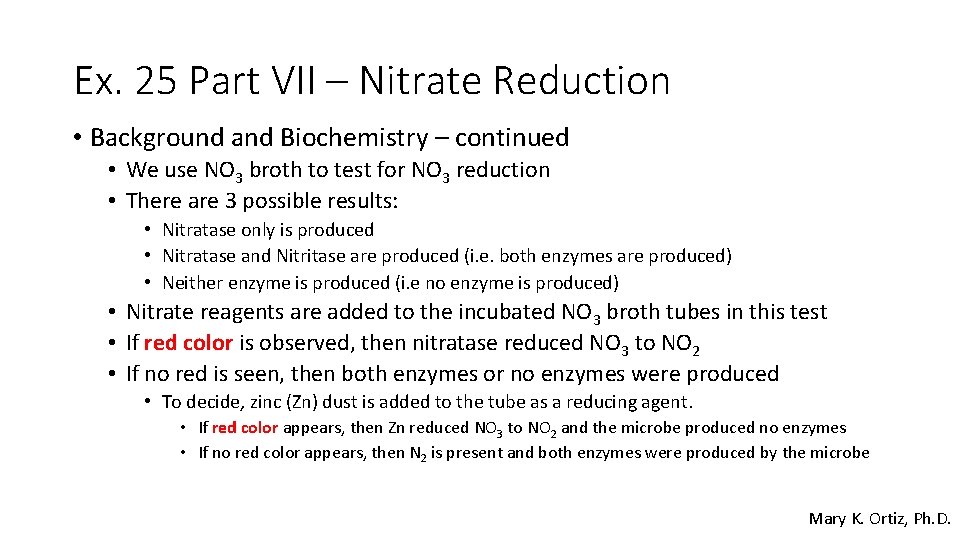
Ex. 25 Part VII – Nitrate Reduction • Background and Biochemistry – continued • We use NO 3 broth to test for NO 3 reduction • There are 3 possible results: • Nitratase only is produced • Nitratase and Nitritase are produced (i. e. both enzymes are produced) • Neither enzyme is produced (i. e no enzyme is produced) • Nitrate reagents are added to the incubated NO 3 broth tubes in this test • If red color is observed, then nitratase reduced NO 3 to NO 2 • If no red is seen, then both enzymes or no enzymes were produced • To decide, zinc (Zn) dust is added to the tube as a reducing agent. • If red color appears, then Zn reduced NO 3 to NO 2 and the microbe produced no enzymes • If no red color appears, then N 2 is present and both enzymes were produced by the microbe Mary K. Ortiz, Ph. D.
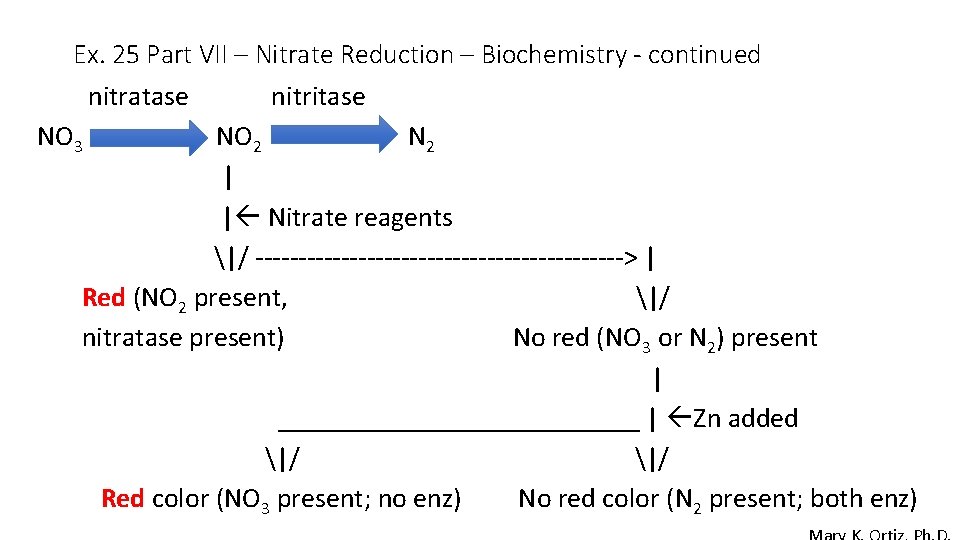
Ex. 25 Part VII – Nitrate Reduction – Biochemistry - continued nitratase NO 3 nitritase NO 2 N 2 | | Nitrate reagents |/ ----------------------> | Red (NO 2 present, |/ nitratase present) No red (NO 3 or N 2) present | _____________ | Zn added |/ Red color (NO 3 present; no enz) No red color (N 2 present; both enz)

Ex. 25 Part VII – Nitrate Reduction Procedure • In this exercise five bacteria are used: • • • E. coli (EC) Proteus vulgaris (PV) Pseudomonas aeruginosa (PA) Staphylococcus epidermidis (SE) Enterococcus faecalis (EF) • Each bacterium is inoculated into a Nitrate broth tube • The tubes are incubated at 37 o. C (98. 6 o. F) for 24 -48 hours • Add the Nitrate reagents to the tubes and observe: • If red color is observed, record results (only nitratase present) • If no red color is observed add Zn • If red color is observed, no enzymes present • If no red color is observed, both enzymes present (ntratase and nitritase present)
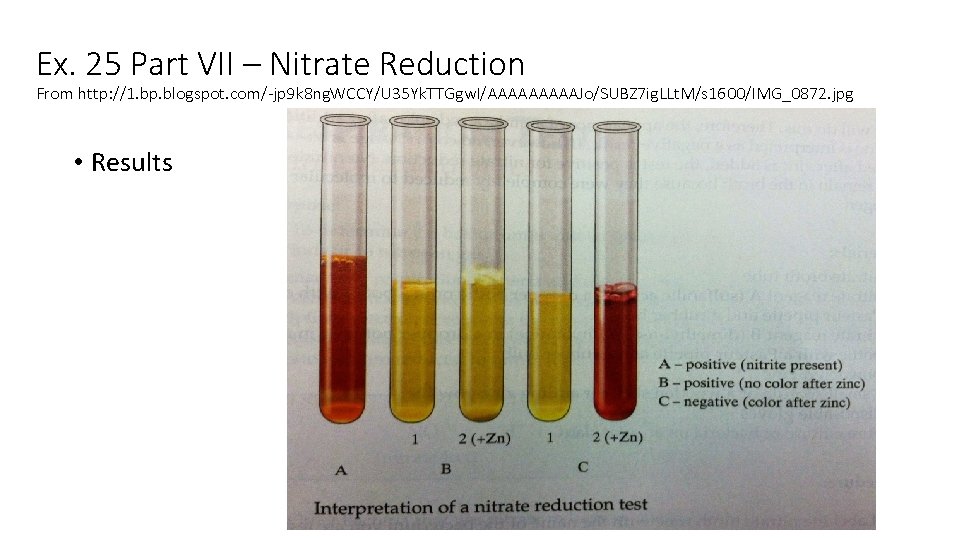
Ex. 25 Part VII – Nitrate Reduction From http: //1. bp. blogspot. com/-jp 9 k 8 ng. WCCY/U 35 Yk. TTGgw. I/AAAAAJo/SUBZ 7 ig. LLt. M/s 1600/IMG_0872. jpg • Results
Ex. 25 Part VII – Nitrate Reduction – Results Bacterium Appearance Interpretation (Result) E. coli Red after Nitrate reagents added + for Nitratase only P. vulgaris Red after Nitrate reagents added P. aeruginosa No red after Nitrate reagents added and no red after Zn added + for both enzymes (Nitratase and Nitritase) S. epidermidis Weak red color after Nitrate reagents added Weakly + for Nitratase only E. faecalis No red after Nitrate reagents added and red after Zn added - for both enzymes (Nitratase and Nitritase) + for Nitratase only

Catalase Testing
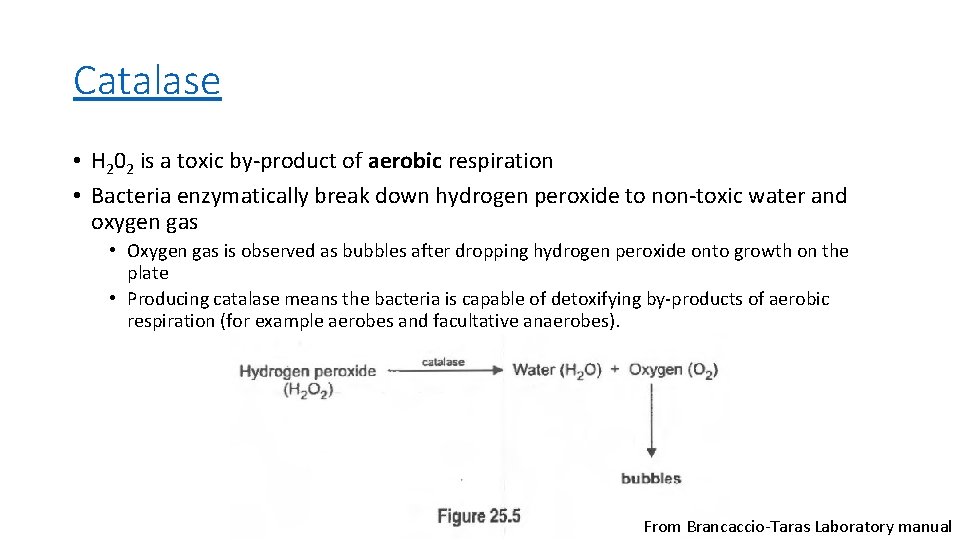
Catalase • H 202 is a toxic by-product of aerobic respiration • Bacteria enzymatically break down hydrogen peroxide to non-toxic water and oxygen gas • Oxygen gas is observed as bubbles after dropping hydrogen peroxide onto growth on the plate • Producing catalase means the bacteria is capable of detoxifying by-products of aerobic respiration (for example aerobes and facultative anaerobes). From Brancaccio-Taras Laboratory manual

Catalase Simulation Positive Result A negative result would not have bubbles. By The original uploader was Philippinjl at French Wikipedia. - Transferred from fr. wikipedia to Commons by Bloodylibu using Commons. Helper. , CC BY-SA 2. 0 fr, https: //commons. wikimedia. org/w/index. php? curid=17397920
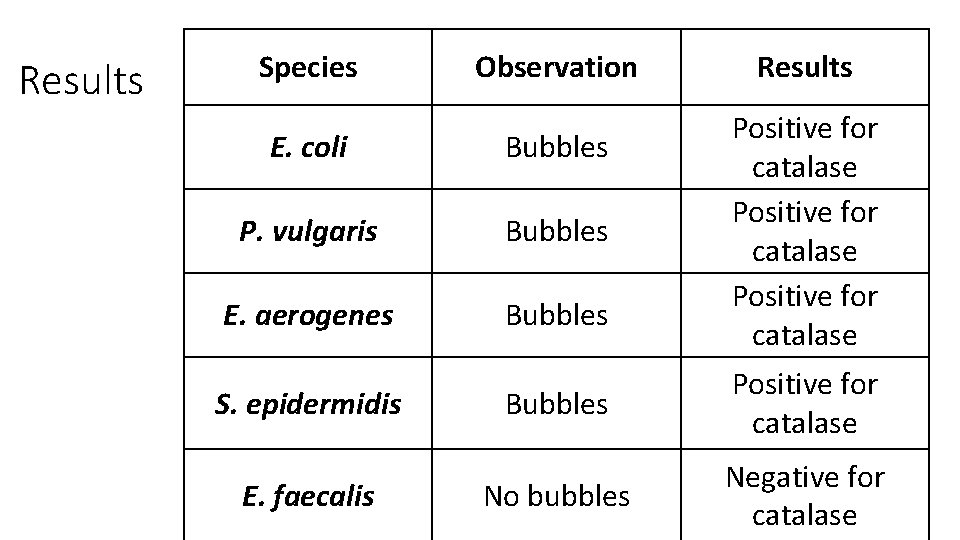
Results Species Observation E. coli Bubbles P. vulgaris Bubbles E. aerogenes Bubbles S. epidermidis E. faecalis Results Positive for catalase Bubbles Positive for catalase No bubbles Negative for catalase
- Slides: 46