Lab 8 PTC Polymerase Chain Reaction Lab Using

Lab 8: PTC Polymerase Chain Reaction Lab Using a Single-Nucleotide Polymorphism (SNP) to Predict Bitter-Tasting Ability

Notebook PTC Polymerase Chain Reaction Lab Objectives • To understand taste perception of the PTC gene and how SNPs can be used as molecular markers to predict phenotypes • To become acquainted with modern molecular genetics techniques used in SNP mapping that include: DNA extraction and purification, polymerase chain reaction (PCR), DNA restriction and gel electrophoresis

PTC Gene (Bitter Tasting Ability)

PTC Gene • Ability to taste PTC (phenylthiocarbamide) is an inherited dominant trait • Varies in the human population and influences taste • PTC taste receptor gene is TAS 2 R 38 (located on chromosome 7) – bitter tasting ability
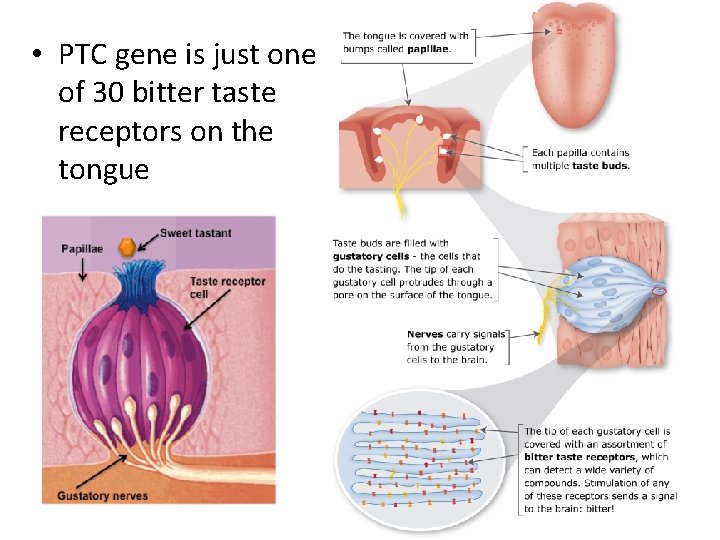
• PTC gene is just one of 30 bitter taste receptors on the tongue

Notebook PTC Procedure A. Collection and Isolation of DNA From Your Cheek Cells B. Amplifying PTC DNA by Polymerase Chain Reaction (PCR) C. Restriction Digest of PCR Products with Hae. III (Restriction Enzyme) D. Gel Electrophoresis of a Restriction Fragment Length Polymorphism (RFLP)
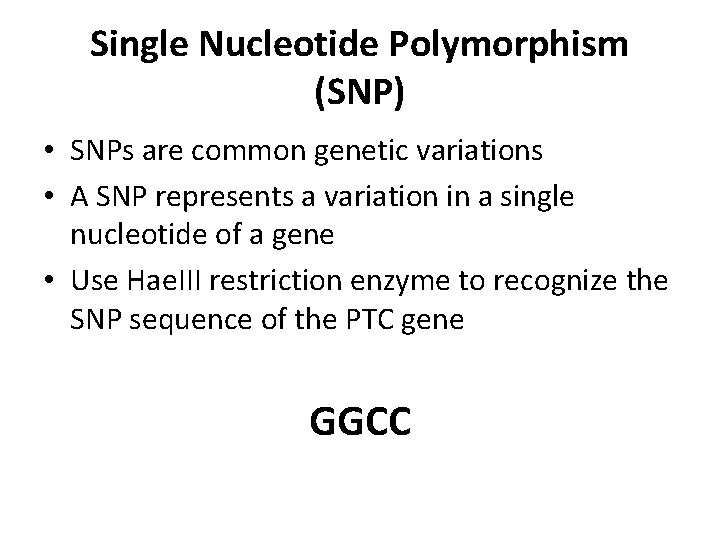
Single Nucleotide Polymorphism (SNP) • SNPs are common genetic variations • A SNP represents a variation in a single nucleotide of a gene • Use Hae. III restriction enzyme to recognize the SNP sequence of the PTC gene GGCC
Single Nucleotide Polymorphism (SNP) • Identify subtle differences in DNA and how they affect you

Notebook Are You a Non-Taster, Weak Taster, or a Strong Taster? Control Paper You Class PTC Non. Taster PTC Weak Taster PTC Strong Taster
Part A: Collection and Isolation of DNA From Your Cheek Cells • • • Swab inside of mouth to obtain cheek cells Rinse cells in saline Centrifuge to obtain the cells in a pellet Use Chelex resin (beads) to extract DNA Heat on a block to melt phospholipids and denature proteins

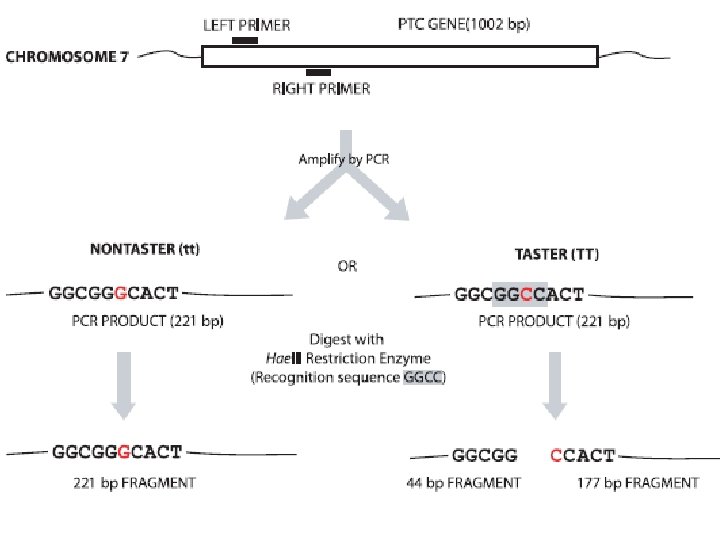
Part B: Amplifying PTC DNA by Polymerase Chain Reaction (PCR) Notebook 1. Add PCR Master Mix (22. 5 u. L) to your DNA (2. 5 u. L) in PCR tube • Gene specific primers – locate the PTC gene • d. NTPs (mixture of A T C G nucleotides) • Salts – neutralize the electrically charged sugar phosphate backbone • p. H of 8. 9 • One Taq DNA Polymerase 2. Perform PCR Amplification using mini. PCR Thermocycler

Amplifying PTC DNA by Polymerase Chain Reaction (PCR)

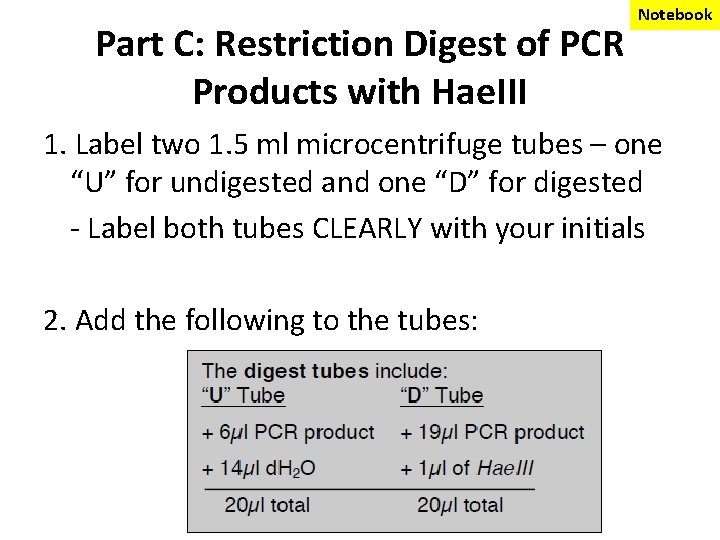
Part C: Restriction Digest of PCR Products with Hae. III Notebook 1. Label two 1. 5 ml microcentrifuge tubes – one “U” for undigested and one “D” for digested - Label both tubes CLEARLY with your initials 2. Add the following to the tubes:

Part C: Restriction Digest of PCR Products with Hae. III (continued) 3. Centrifuge the “D” tube to pool reagents 4. Place “D” tube in a 37°C water bath for 15 minutes 5. Store in freezer until gel electrophoresis experiment Notebook
Notebook Electrophoresis Prediction • Based on the results of the PTC taste test (phenotype), draw your predicted electrophoresis results • Label each band on the gel with the appropriate fragment size in base pairs

Notebook Part D: Gel Electrophoresis of a Restriction Fragment Length Polymorphism (RFLP) 1. Add 2 µL of loading dye to the “U” tube and the “D” tube. 2. Using a p 20 micropipette, load 18 µL of each sample into each well. 18 µL of DNA ladder will also be loaded into a well. 3. Record the well number(s) you loaded your samples into the gel. 4. Run the gel at 100 volts for 30 minutes and observe gel under UV light. Take a picture of the final gel.
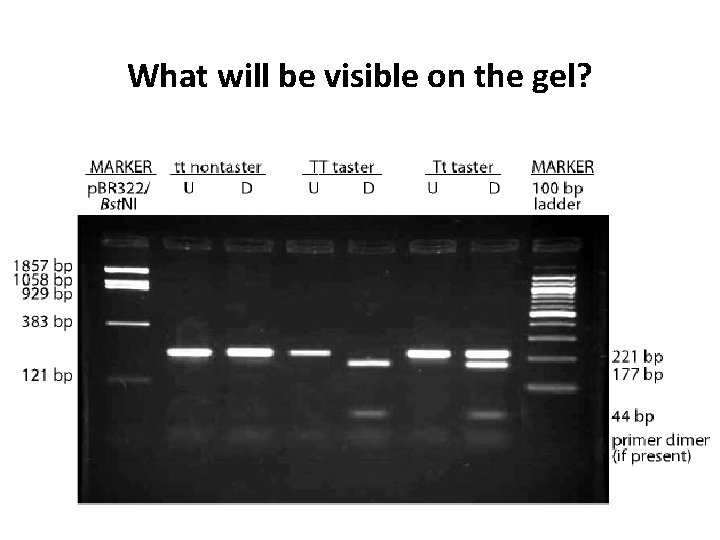
What will be visible on the gel?
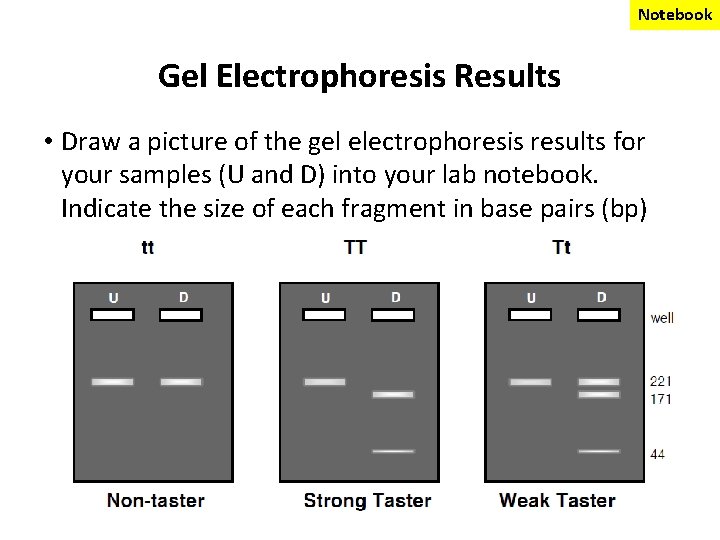
Notebook Gel Electrophoresis Results • Draw a picture of the gel electrophoresis results for your samples (U and D) into your lab notebook. Indicate the size of each fragment in base pairs (bp)
- Slides: 20