Lab 6 Isolation Techniques DNA Isolation from Haman
Lab 6 Isolation Techniques

DNA Isolation from Haman Blood Cells Objective: • Isolating DNA molecules from white blood cells (WBCs) and visualizing them directly. • Blood is composed of liquid plasma and solid substances including blood cells. • WBCs have nuclei that contains the nucleic acids. 1 -Cellular membrane and nucleic membrane must be destroyed first 2 - Other components of the cell like proteins must be excluded 3 -finally DNA molecules can be collected and directly visualized.

In this experiment : 1 - cellular and nucleic membranes are lysed with using lysis buffer 2 - Proteins are precipitated by using phenol and chloroform. 3 - Protect DNA from lysis by DNase enzyme by using EDTA which inhibit DNase 4 - Collect DNA molecules by adding both sodium acetate and isopropanol. Reagents : Chemicals such as 1 -EDTA (Ethylene diamine tetra acetate) which removes Mg+2 ions that is essential for preserving the overall structure of the cell membrane. 2 -SDS (Sodium dodecyl sulfate), which aids in disrupting the cell membranes by removing the lipids of the cell membranes, are included in the extraction. 3 -buffer: which lysing the cells, the final step in the preparation of a cell extract is removal of insoluble cell debris

• Purification of DNA from cell extract: • In addition to DNA the cell extract will contain significant quantities of protein and RNA. A variety of procedures can be used to remove these contaminants, leaving the DNA in a pure form. Reagents Phenol: • The standard way to de-proteinize a cell extract is to add phenol or a 1: 1 mixture of phenol: chloroform. These organic solvents precipitate proteins but leave the nucleic acids in aqueous solutions. The aqueous solution of nucleic acid can be removed with a pipette. Ribonuclease enzyme: • Ribonuclease enzyme remove RNA by degrade these molecules into ribonucleotide subunits.

Materials: - Lysis buffer - SE buffer -Blood with anti-coagulant - Proteinase K (10 mg/ml) - SDS reagent (contains detergent and EDTA) - Chloroform/ isoamyl alcohol 24: 1 - Sodium acetate - Isopropanol - Micropipettes and tips - Refrigerated centrifuge and 15 ml Eppendorf tubes
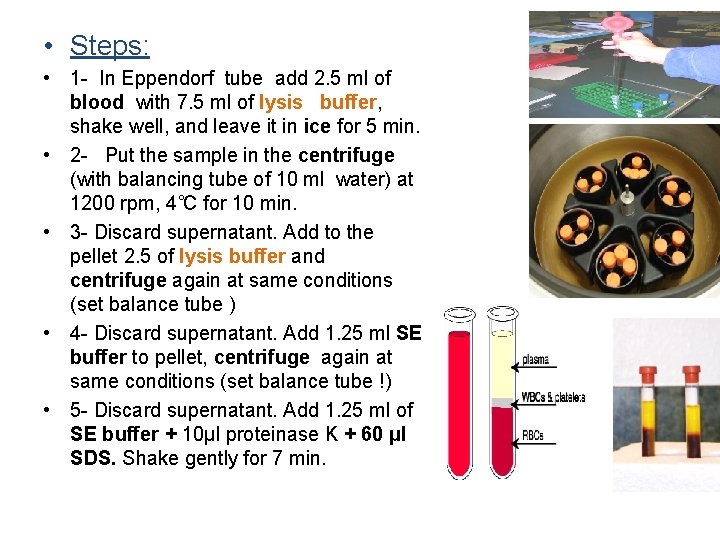
• Steps: • 1 - In Eppendorf tube add 2. 5 ml of blood with 7. 5 ml of lysis buffer, shake well, and leave it in ice for 5 min. • 2 - Put the sample in the centrifuge (with balancing tube of 10 ml water) at 1200 rpm, 4˚C for 10 min. • 3 - Discard supernatant. Add to the pellet 2. 5 of lysis buffer and centrifuge again at same conditions (set balance tube ) • 4 - Discard supernatant. Add 1. 25 ml SE buffer to pellet, centrifuge again at same conditions (set balance tube !) • 5 - Discard supernatant. Add 1. 25 ml of SE buffer + 10μl proteinase K + 60 μl SDS. Shake gently for 7 min.

6 - Add 1. 25 ml SE buffer + 2. 5 ml phenol. Shake well for 5 min then centrifuge again at 3000 rpm, 10 ˚C for 5 min. 7 - Transfer supernatant to new tube. Add 2. 5 ml Phenol/ chloroform/isoamy alcohol. Shake well for 5 min, then centrifuge Again at same conditions. 8 - Transfer supernatant to new tube. Add 2. 5 ml chloroform/isoamyl alcohol. Shake well for 5 min, then centrifuge again at same conditions. 9 - Transfer supernatant to new tube. Add 75 μl 3 M Sodium acetate + 2. 5 ml isopropanol. Observe DNA fibers visually. vortex shaker

DNA Isolation from Blood Animation • http: //www. iupui. edu/~wellsctr/MMIA/isolating_dna/dna_isolation_re v. swf
- Slides: 8