KASP Workshop Science for a safer world Overview



![Acceptable submission formats Acceptable sequence submission formats Example [First allele/second allele] ATAACTACTTTTAAAGGCACATTATTCAACCTCACTGTGCATT TCATCCTC[A/T]TGCATAATGACAGTTATTCTCTCCCAAGTCT CCTTCTGGGACAGTTAAAG Acceptable submission formats Acceptable sequence submission formats Example [First allele/second allele] ATAACTACTTTTAAAGGCACATTATTCAACCTCACTGTGCATT TCATCCTC[A/T]TGCATAATGACAGTTATTCTCTCCCAAGTCT CCTTCTGGGACAGTTAAAG](https://slidetodoc.com/presentation_image_h/a14a6c0a3c84360a27e93613bfdddca8/image-6.jpg)











- Slides: 53

KASP Workshop Science for a safer world

Overview of topics to be covered • • Sequence submission Running KASP genotyping reactions Different Platforms Troubleshooting KASP data- where support is available • Design solutions

Sequence submission − how to submit sequence information for KASP assay design

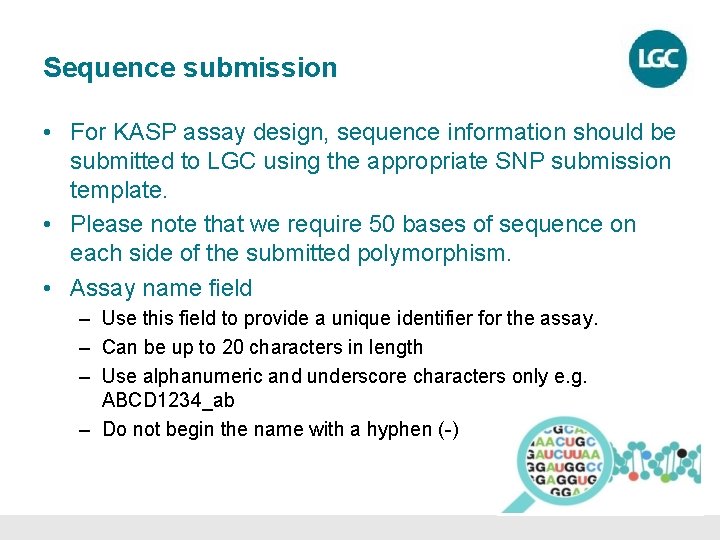
Sequence submission • For KASP assay design, sequence information should be submitted to LGC using the appropriate SNP submission template. • Please note that we require 50 bases of sequence on each side of the submitted polymorphism. • Assay name field – Use this field to provide a unique identifier for the assay. – Can be up to 20 characters in length – Use alphanumeric and underscore characters only e. g. ABCD 1234_ab – Do not begin the name with a hyphen (-)

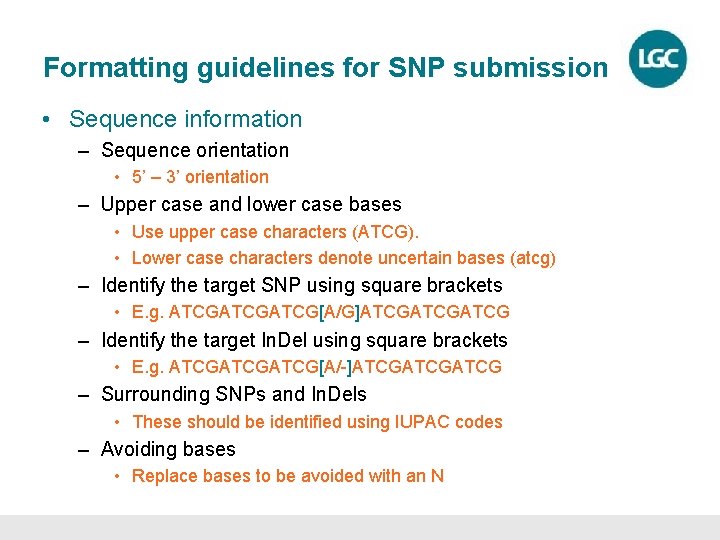
Formatting guidelines for SNP submission • Sequence information – Sequence orientation • 5’ – 3’ orientation – Upper case and lower case bases • Use upper case characters (ATCG). • Lower case characters denote uncertain bases (atcg) – Identify the target SNP using square brackets • E. g. ATCGATCG[A/G]ATCGATCG – Identify the target In. Del using square brackets • E. g. ATCGATCG[A/-]ATCGATCG – Surrounding SNPs and In. Dels • These should be identified using IUPAC codes – Avoiding bases • Replace bases to be avoided with an N
![Acceptable submission formats Acceptable sequence submission formats Example First allelesecond allele ATAACTACTTTTAAAGGCACATTATTCAACCTCACTGTGCATT TCATCCTCATTGCATAATGACAGTTATTCTCTCCCAAGTCT CCTTCTGGGACAGTTAAAG Acceptable submission formats Acceptable sequence submission formats Example [First allele/second allele] ATAACTACTTTTAAAGGCACATTATTCAACCTCACTGTGCATT TCATCCTC[A/T]TGCATAATGACAGTTATTCTCTCCCAAGTCT CCTTCTGGGACAGTTAAAG](https://slidetodoc.com/presentation_image_h/a14a6c0a3c84360a27e93613bfdddca8/image-6.jpg)
Acceptable submission formats Acceptable sequence submission formats Example [First allele/second allele] ATAACTACTTTTAAAGGCACATTATTCAACCTCACTGTGCATT TCATCCTC[A/T]TGCATAATGACAGTTATTCTCTCCCAAGTCT CCTTCTGGGACAGTTAAAG IUPAC code ATAACTACTTTTAAAGGCACATTATTCAACCTCACTGTGCATT TCATCCTC[W]TGCATAATGACAGTTATTCTCTCCCAAGTCTC CTTCTGGGACAGTTAAAG Rs Number Rs 123456* *N. B. It is advisable to submit the sequence in addition to the Rs number to ensure that LGC Genomics are using exactly the sequence that you require. Indels ATAACTACTTTTAAAGGCACATTATTCAACCTCACTGTGCATT TCATCCTC[/T]TGCATAATGACAGTTATTCTCTCCCAAGTCTC CTTCTGGGACAGTTAAAG

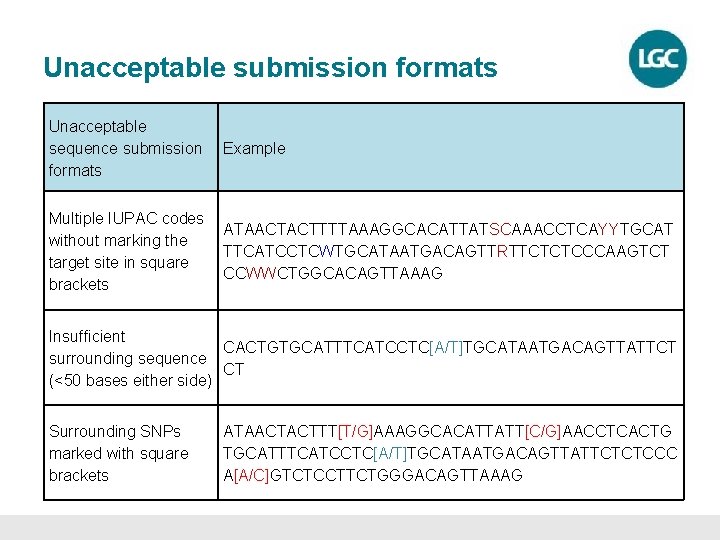
Unacceptable submission formats Unacceptable sequence submission formats Example Multiple IUPAC codes without marking the target site in square brackets ATAACTACTTTTAAAGGCACATTATSCAAACCTCAYYTGCAT TTCATCCTCWTGCATAATGACAGTTRTTCTCTCCCAAGTCT CCWWCTGGCACAGTTAAAG Insufficient CACTGTGCATTTCATCCTC[A/T]TGCATAATGACAGTTATTCT surrounding sequence CT (<50 bases either side) Surrounding SNPs marked with square brackets ATAACTACTTT[T/G]AAAGGCACATTATT[C/G]AACCTCACTG TGCATTTCATCCTC[A/T]TGCATAATGACAGTTATTCTCTCCC A[A/C]GTCTCCTTCTGGGACAGTTAAAG

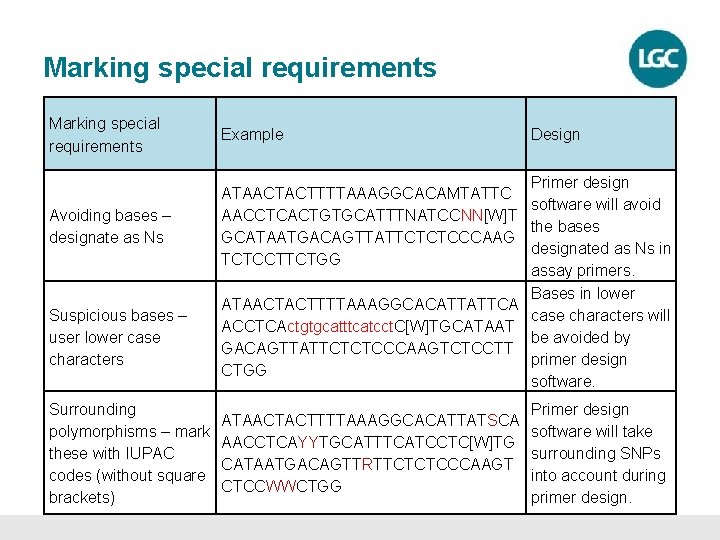
Marking special requirements Avoiding bases – designate as Ns Suspicious bases – user lower case characters Surrounding polymorphisms – mark these with IUPAC codes (without square brackets) Example Design Primer design ATAACTACTTTTAAAGGCACAMTATTC software will avoid AACCTCACTGTGCATTTNATCCNN[W]T the bases GCATAATGACAGTTATTCTCTCCCAAG designated as Ns in TCTCCTTCTGG assay primers. Bases in lower ATAACTACTTTTAAAGGCACATTATTCA case characters will ACCTCActgtgcatttcatcct. C[W]TGCATAAT be avoided by GACAGTTATTCTCTCCCAAGTCTCCTT primer design CTGG software. Primer design ATAACTACTTTTAAAGGCACATTATSCA software will take AACCTCAYYTGCATTTCATCCTC[W]TG surrounding SNPs CATAATGACAGTTRTTCTCTCCCAAGT into account during CTCCWWCTGG primer design.

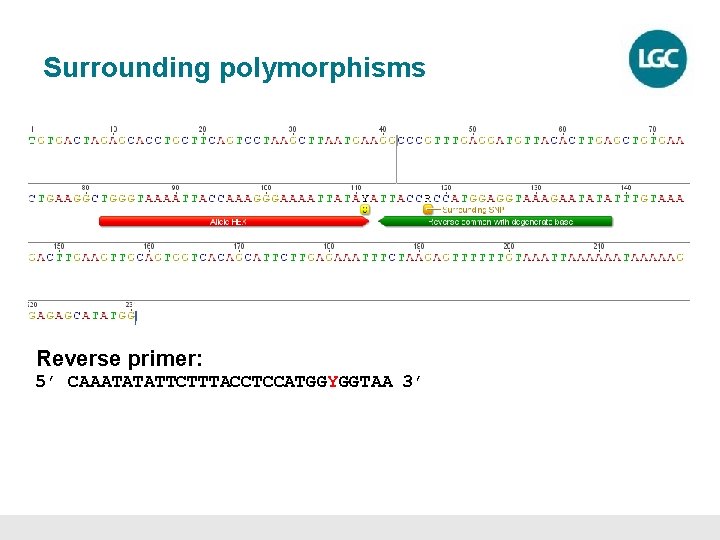
Surrounding polymorphisms Reverse primer: 5’ CAAATATATTCTTTACCTCCATGGYGGTAA 3’

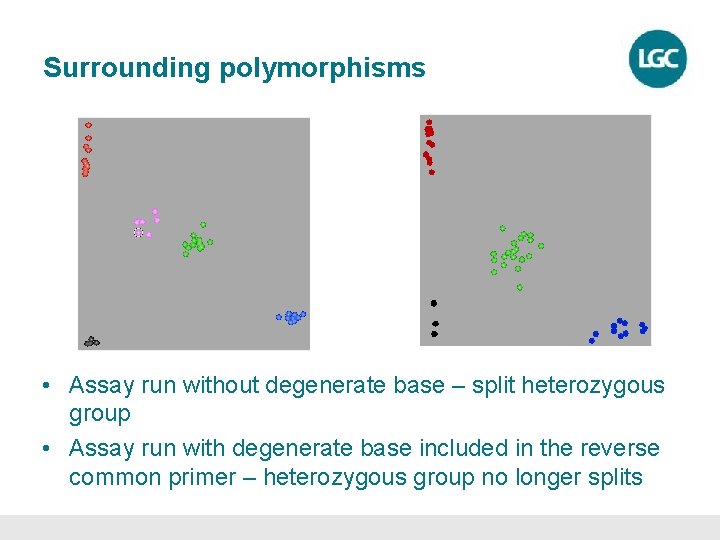
Surrounding polymorphisms • Assay run without degenerate base – split heterozygous group • Assay run with degenerate base included in the reverse common primer – heterozygous group no longer splits

Three adjacent SNPs • If the region of interest has 3 adjacent SNPs, three KASP assays will be required XXXXX [C/A][A/G][G/T] XXXXX • For each assay – submit the SNP of interest in square brackets – submit the surrounding SNPs using IUPAC codes SNP 1 SNP 2 SNP 3 XXXXX [C/A] R K XXXXX M [A/G] K XXXXX M R [G/T] XXXXX

Running KASP genotyping reactions − How to set-up and run KASP genotyping reactions

Customer requirements • FRET-capable plate reader • PCR microtitre plate (96 -, 384 - or 1536 -well) • DNA samples at an appropriate concentration for KASP reactions – Minimum 22 DNA samples per assay • PCR grade water – Run 2 NTC wells per assay • Optically clear seal

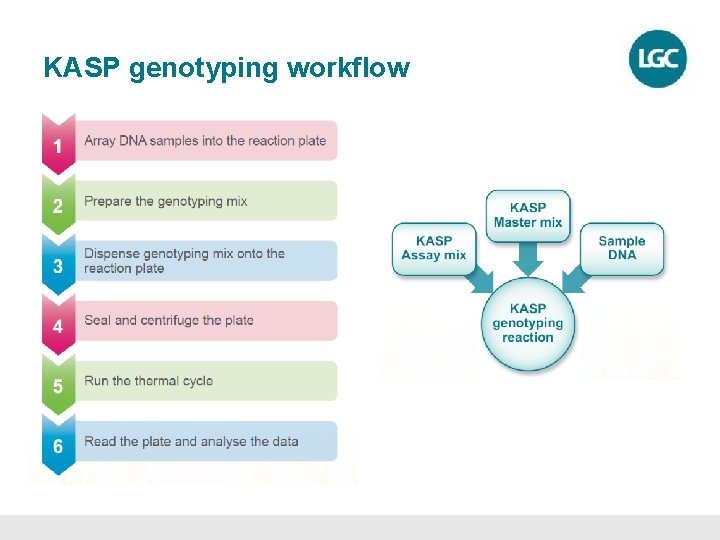
KASP genotyping workflow

Setting up a KASP genotyping reaction • DNA samples at the appropriate concentration for KASP – 50 ng per reaction (human genome size) • 96 - or 384 -well PCR plates • Arrayed DNA can be dried down or stored in hydrated form

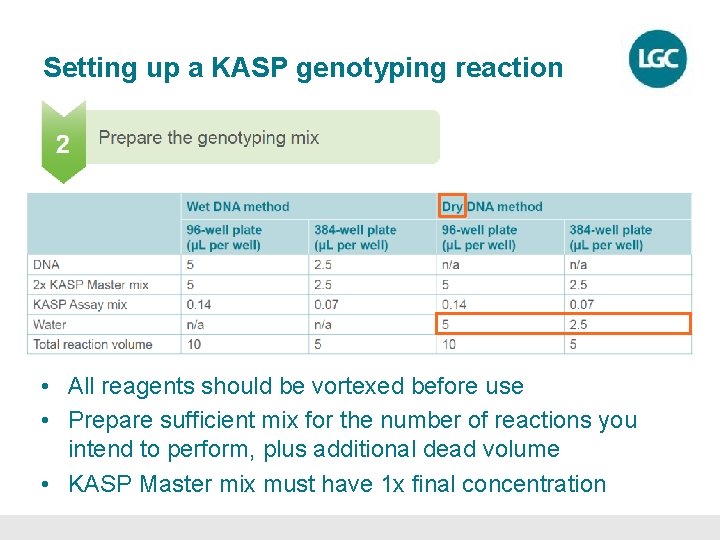
Setting up a KASP genotyping reaction • All reagents should be vortexed before use • Prepare sufficient mix for the number of reactions you intend to perform, plus additional dead volume • KASP Master mix must have 1 x final concentration

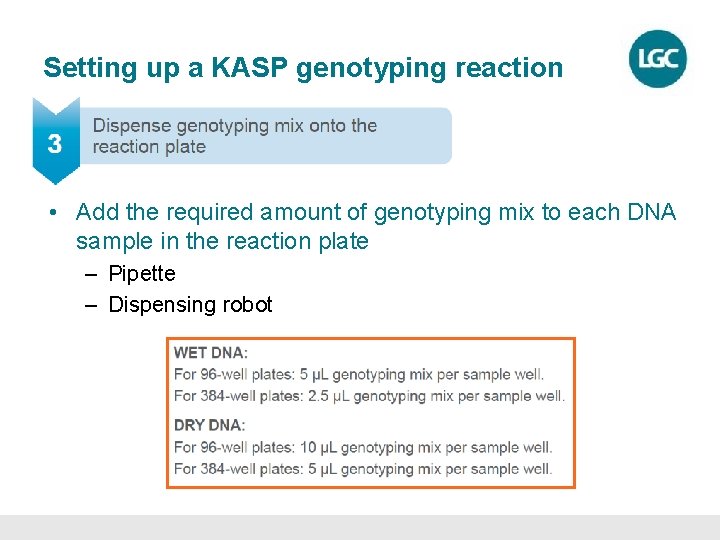
Setting up a KASP genotyping reaction • Add the required amount of genotyping mix to each DNA sample in the reaction plate – Pipette – Dispensing robot

Setting up a KASP genotyping reaction • Seal the plate with an optically clear seal • Centrifuge the plate briefly to ensure all reaction volume is at the bottom of the wells

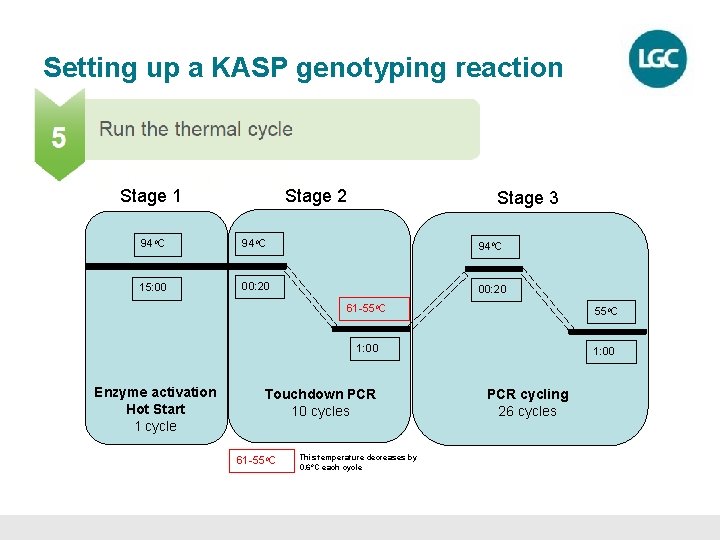
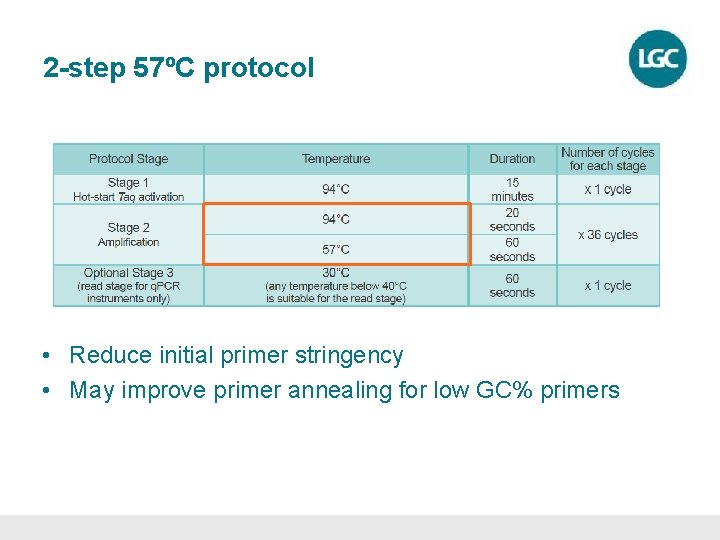
Setting up a KASP genotyping reaction • KASP chemistry can be used with any standard thermal cycler • The KASP thermal cycle is a two-step PCR program – Denaturation – Annealing and extension – all in one step. • 10 cycles of touchdown PCR – Gradually decrease the annealing temperature each cycle to increase specificity of the reaction. • 26 cycles of standard PCR

Setting up a KASP genotyping reaction Stage 1 Stage 2 Stage 3 94 o. C 15: 00 00: 20 Enzyme activation Hot Start 1 cycle 61 -55 o. C 1: 00 Touchdown PCR 10 cycles 61 -55 o. C This temperature decreases by 0. 6 o. C each cycle PCR cycling 26 cycles

Setting up a KASP genotyping reaction • Read the plate in a FRET-capable plate reader • All plates should be read below 40ºC • To analyse the data, import it into a genotype cluster analysis software package • Data can be plotted and genotyping clusters analysed

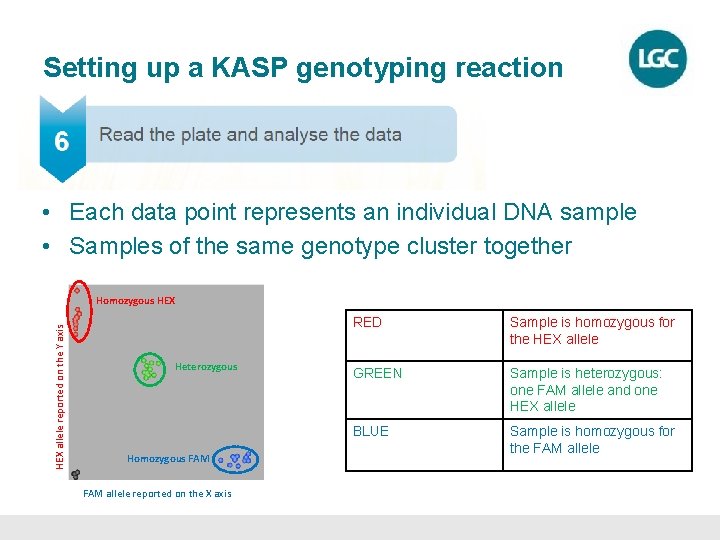
Setting up a KASP genotyping reaction • Each data point represents an individual DNA sample • Samples of the same genotype cluster together HEX allele reported on the Y axis Homozygous HEX Heterozygous Homozygous FAM allele reported on the X axis RED Sample is homozygous for the HEX allele GREEN Sample is heterozygous: one FAM allele and one HEX allele BLUE Sample is homozygous for the FAM allele

Recycling • Efficiency of KASP dependent on a number of factors – DNA concentration – Composition of DNA sequence surrounding polymorphism • Different KASP assays will reach completion at different rates • Can require additional cycles to product clear genotyping clusters • Reaction plate should be thermally cycled further – “Recycling”

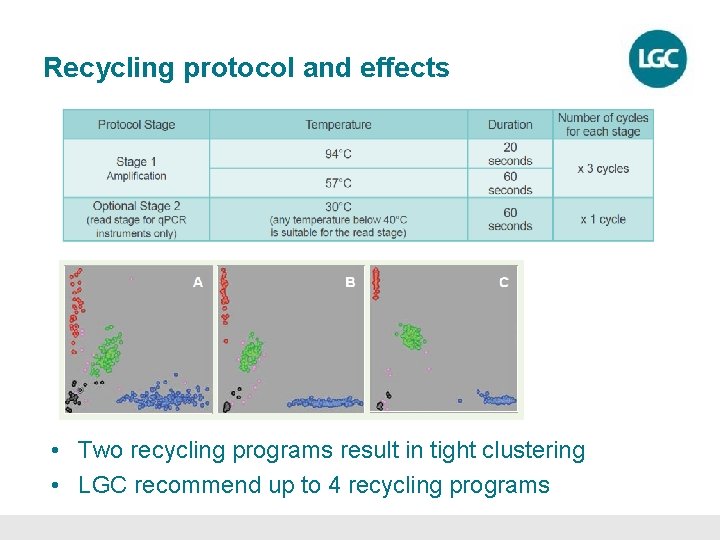
Recycling protocol and effects • Two recycling programs result in tight clustering • LGC recommend up to 4 recycling programs

Thermal cycling conditions • Standard KASP thermal cycling protocol – 61 -55ºC touchdown protocol (36 cycles) • If recycling improves clustering, add cycles to initial thermal cycle e. g. run 39 cycles each time instead • Alternative KASP thermal cycling protocols – 68 -62ºC touchdown protocol • Annealing temperatures >65ºC (high GC%) – 2 -step 57ºC protocol • Annealing temperatures <60ºC (low GC%)

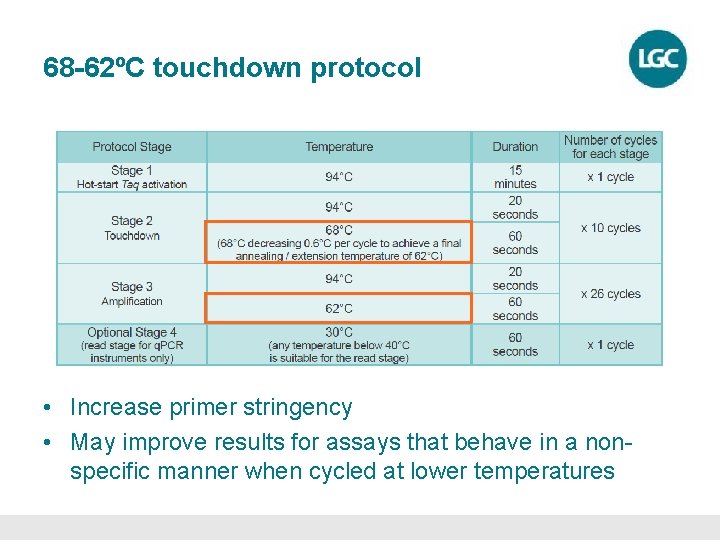
68 -62ºC touchdown protocol • Increase primer stringency • May improve results for assays that behave in a nonspecific manner when cycled at lower temperatures

2 -step 57ºC protocol • Reduce initial primer stringency • May improve primer annealing for low GC% primers

Platforms − The range of platforms that KASP can be run on

Running KASP on q. PCR instruments ABI: 7000 -series, Viia 7, Quant Studio-series Roche: LC 480 v. I and v. II, LC 2. 0 & LC 96 BIO-RAD: CFX, i. Cycler i. Q, Myi. Q & Myi. Q 2 Thermo Scientific: Piko. Real Illumina: Eco RT-PCR Agilent: Mx 3000 / 3500 P & Mx 4000 P 29

Manuals available and coming soon Currently available: • • • ABI 7900 ABI 7500 ABI Viia 7 ABI Step. One and Step. One Plus BIORAD CFX Agilent Mx-series Illumina Eco. RT Roche LC 480 (v. I and v. II) Workaround document for LC 96 Coming soon: • Fluidigm (updated)

Troubleshooting KASP data − How to troubleshoot data and improve results

Common causes of unexpected results • • • Storage – aliquot, store in dark (Master mix only) Thaw fully before use Mix at every stage DNA concentration DNA quality Thermal cycle correctly programmed Accurate pipetting KASP Master mix – appropriate ROX level Plates sealed sufficiently Plate read below 40ºC Correct excitation and emission values

Common assay optimisations • Recycling to reach reaction completion • Altering thermal cycling conditions – 68 -62ºC touchdown protocol – 2 -step 57ºC protocol • Altering the magnesium concentration of the reactions – Target SNPs in low GC% regions may require more magnesium – Increase incrementally ~0. 3 m. M • Adding DMSO – Target SNPs in high GC% regions may benefit from addition of DMSO – Add 5 -10% DMSO to the final volume of the reaction – Use standard magnesium concentration

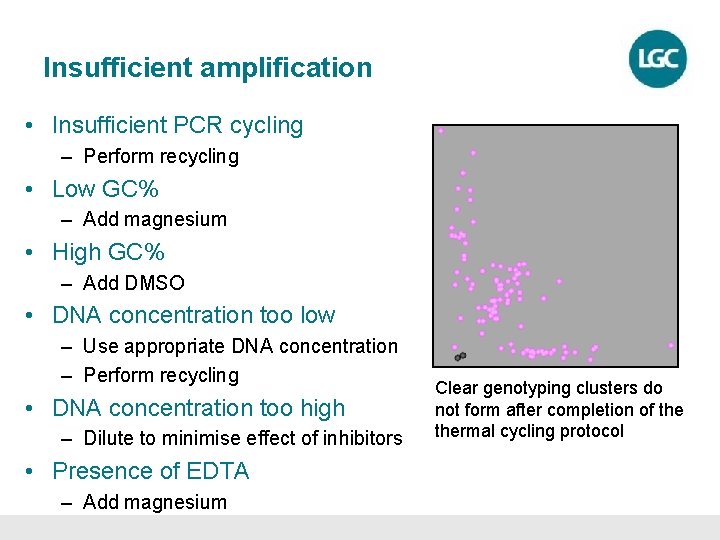
Insufficient amplification • Insufficient PCR cycling – Perform recycling • Low GC% – Add magnesium • High GC% – Add DMSO • DNA concentration too low – Use appropriate DNA concentration – Perform recycling • DNA concentration too high – Dilute to minimise effect of inhibitors • Presence of EDTA – Add magnesium Clear genotyping clusters do not form after completion of thermal cycling protocol

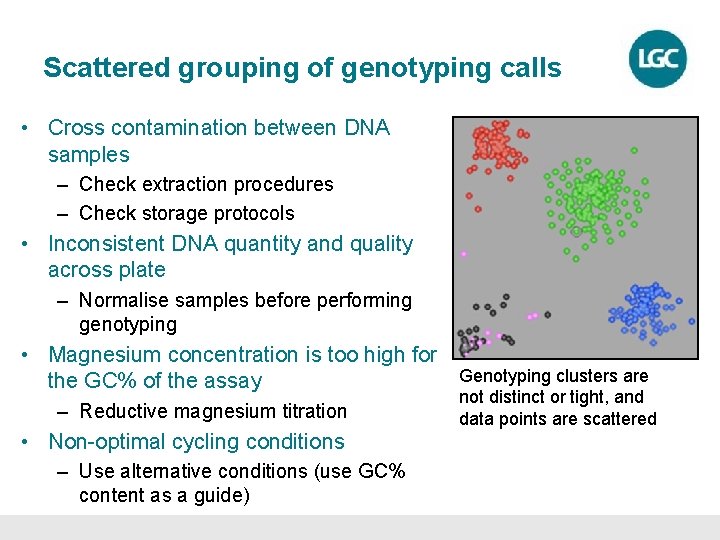
Scattered grouping of genotyping calls • Cross contamination between DNA samples – Check extraction procedures – Check storage protocols • Inconsistent DNA quantity and quality across plate – Normalise samples before performing genotyping • Magnesium concentration is too high for the GC% of the assay – Reductive magnesium titration • Non-optimal cycling conditions – Use alternative conditions (use GC% content as a guide) Genotyping clusters are not distinct or tight, and data points are scattered

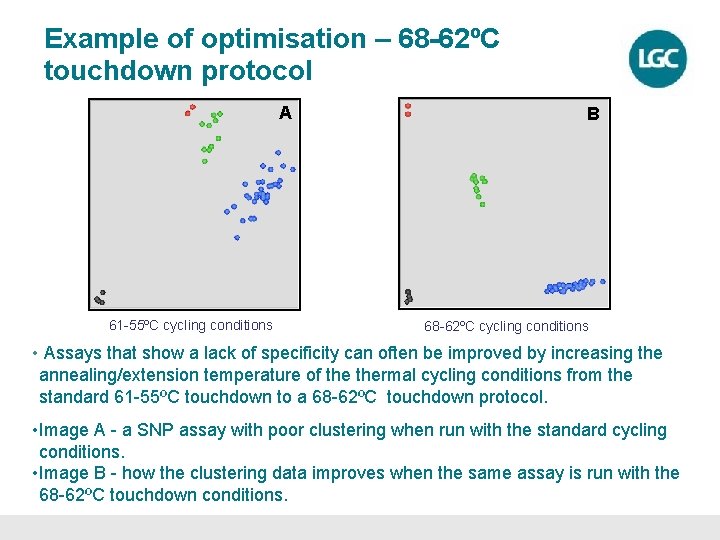
Example of optimisation – 68 -62ºC touchdown protocol A 61 -55ºC cycling conditions B 68 -62ºC cycling conditions • Assays that show a lack of specificity can often be improved by increasing the annealing/extension temperature of thermal cycling conditions from the standard 61 -55ºC touchdown to a 68 -62ºC touchdown protocol. • Image A - a SNP assay with poor clustering when run with the standard cycling conditions. • Image B - how the clustering data improves when the same assay is run with the 68 -62ºC touchdown conditions.

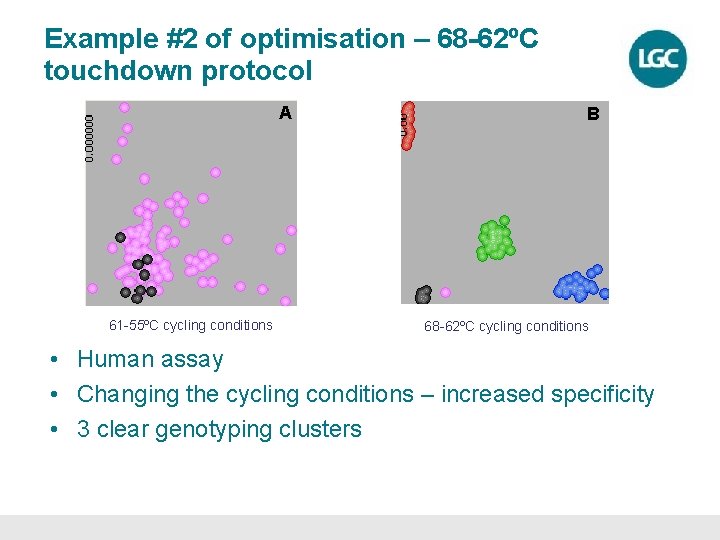
Example #2 of optimisation – 68 -62ºC touchdown protocol A 61 -55ºC cycling conditions B 68 -62ºC cycling conditions • Human assay • Changing the cycling conditions – increased specificity • 3 clear genotyping clusters

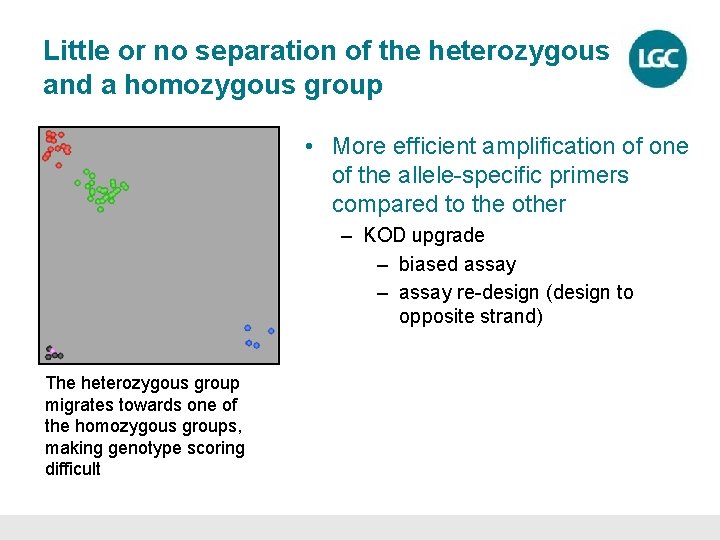
Little or no separation of the heterozygous and a homozygous group • More efficient amplification of one of the allele-specific primers compared to the other – KOD upgrade – biased assay – assay re-design (design to opposite strand) The heterozygous group migrates towards one of the homozygous groups, making genotype scoring difficult

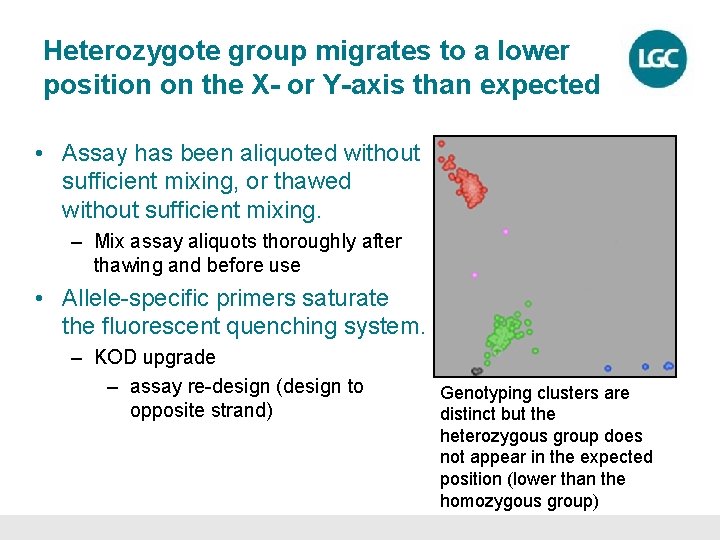
Heterozygote group migrates to a lower position on the X- or Y-axis than expected • Assay has been aliquoted without sufficient mixing, or thawed without sufficient mixing. – Mix assay aliquots thoroughly after thawing and before use • Allele-specific primers saturate the fluorescent quenching system. – KOD upgrade – assay re-design (design to opposite strand) Genotyping clusters are distinct but the heterozygous group does not appear in the expected position (lower than the homozygous group)

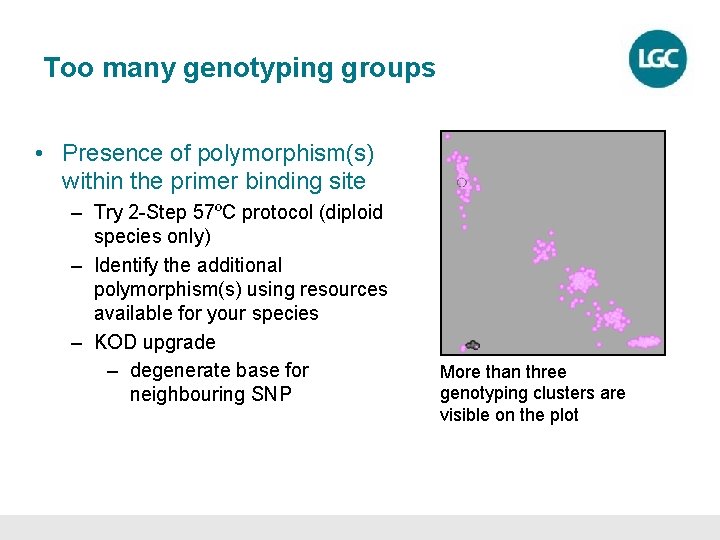
Too many genotyping groups • Presence of polymorphism(s) within the primer binding site – Try 2 -Step 57ºC protocol (diploid species only) – Identify the additional polymorphism(s) using resources available for your species – KOD upgrade – degenerate base for neighbouring SNP More than three genotyping clusters are visible on the plot

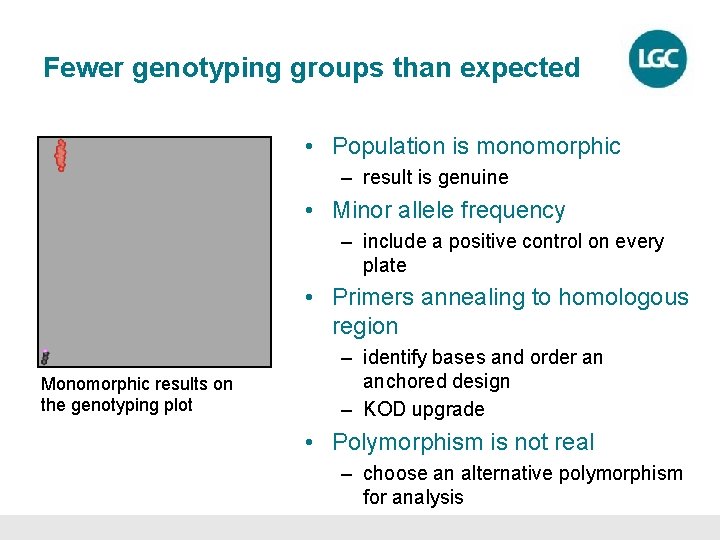
Fewer genotyping groups than expected • Population is monomorphic – result is genuine • Minor allele frequency – include a positive control on every plate • Primers annealing to homologous region Monomorphic results on the genotyping plot – identify bases and order an anchored design – KOD upgrade • Polymorphism is not real – choose an alternative polymorphism for analysis

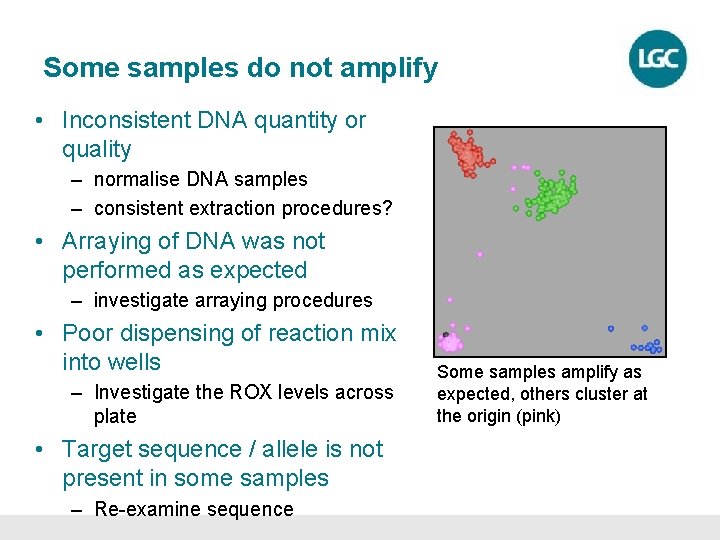
Some samples do not amplify • Inconsistent DNA quantity or quality – normalise DNA samples – consistent extraction procedures? • Arraying of DNA was not performed as expected – investigate arraying procedures • Poor dispensing of reaction mix into wells – Investigate the ROX levels across plate • Target sequence / allele is not present in some samples – Re-examine sequence Some samples amplify as expected, others cluster at the origin (pink)

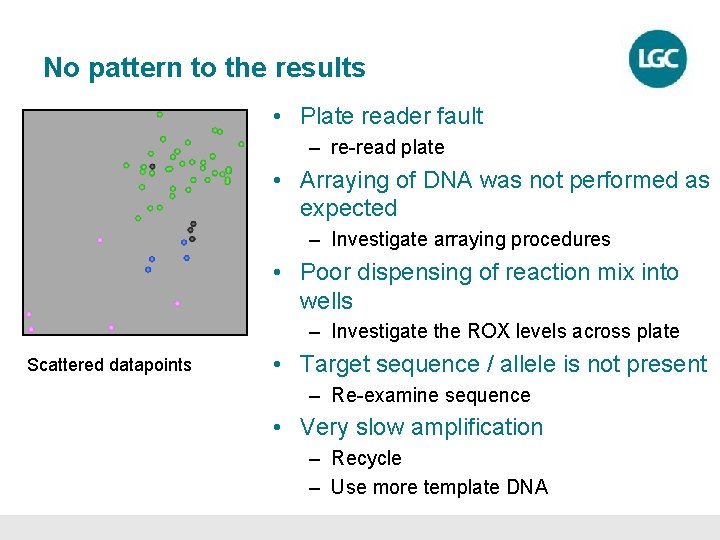
No pattern to the results • Plate reader fault – re-read plate • Arraying of DNA was not performed as expected – Investigate arraying procedures • Poor dispensing of reaction mix into wells – Investigate the ROX levels across plate Scattered datapoints • Target sequence / allele is not present – Re-examine sequence • Very slow amplification – Recycle – Use more template DNA

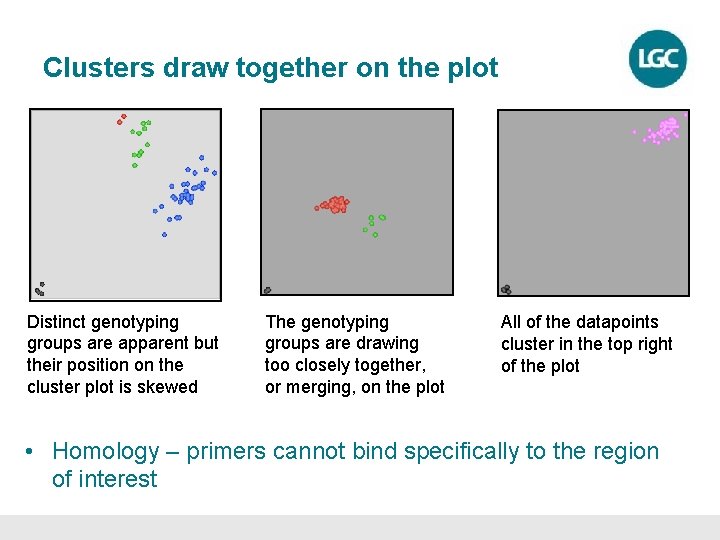
Clusters draw together on the plot Distinct genotyping groups are apparent but their position on the cluster plot is skewed The genotyping groups are drawing too closely together, or merging, on the plot All of the datapoints cluster in the top right of the plot • Homology – primers cannot bind specifically to the region of interest

Design Solutions - Dealing with homology 45

Homology • Primers are annealing to identical homologous regions • Allele-specific primers are not able to bind specifically to their target template – experimentally increase the annealing temperature to force improved specificity (annealing stringency) – Add 5% DMSO – may work – KOD upgrade – anchored primers – perform a BLAST, identify unique bases

Assay design solutions • KASP is great for finding solutions for polyploid organisms • Similar solutions can be used for regions of high identity or pseudogenes within genome. • Assays can be designed within highly polymorphic regions by adaption of primers. • Large In. Del region can be screened for. • KASP is PCR so can be optimised in similar ways to increase the AT / GC range

Solutions for homology and polyploid organisms. • Solutions to tricky problems can often be found for some assays by anchoring • More solutions come from having the correct sequence giving more options for designing better primers. • Primers with degeneracy can be used to avoid associated problem of highly polymorphic regions • Designing in either orientation is often a very easy solution for genomes that are not well annotated Chicken is a very good example and works well.

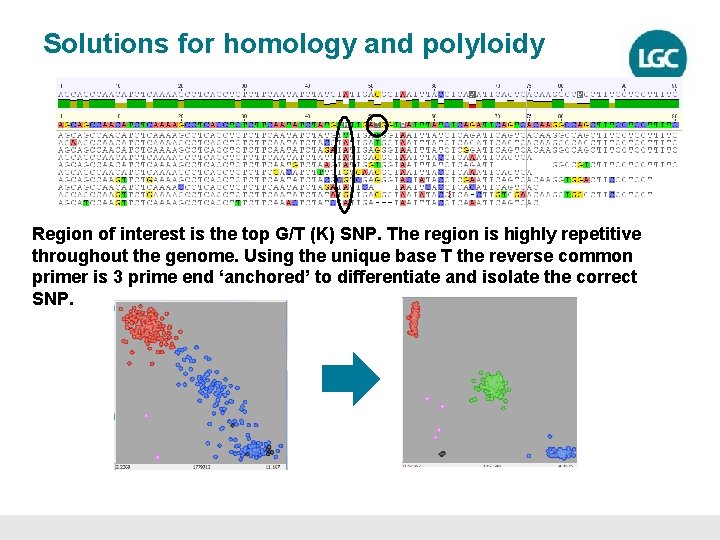
Solutions for homology and polyloidy Region of interest is the top G/T (K) SNP. The region is highly repetitive throughout the genome. Using the unique base T the reverse common primer is 3 prime end ‘anchored’ to differentiate and isolate the correct SNP.

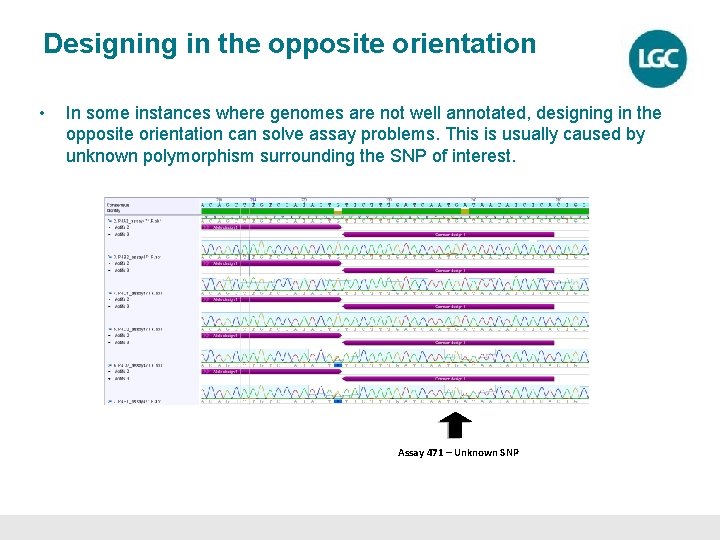
Designing in the opposite orientation • In some instances where genomes are not well annotated, designing in the opposite orientation can solve assay problems. This is usually caused by unknown polymorphism surrounding the SNP of interest. Assay 471 – Unknown SNP

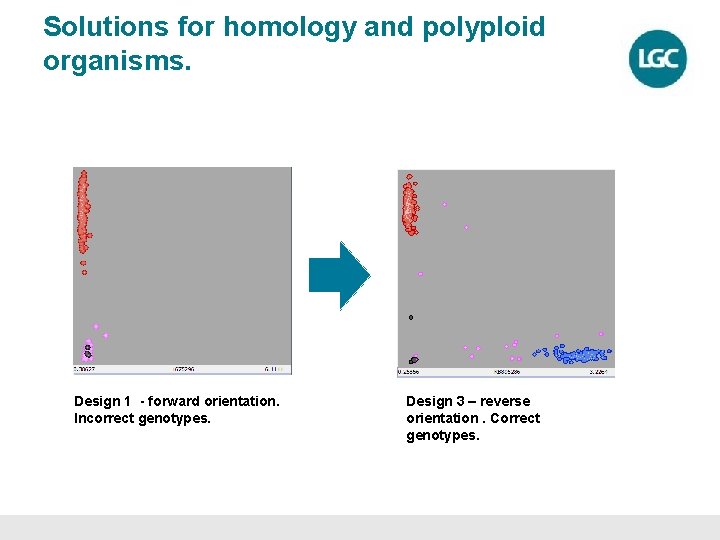
Solutions for homology and polyploid organisms. Design 1 - forward orientation. Incorrect genotypes. Design 3 – reverse orientation. Correct genotypes.

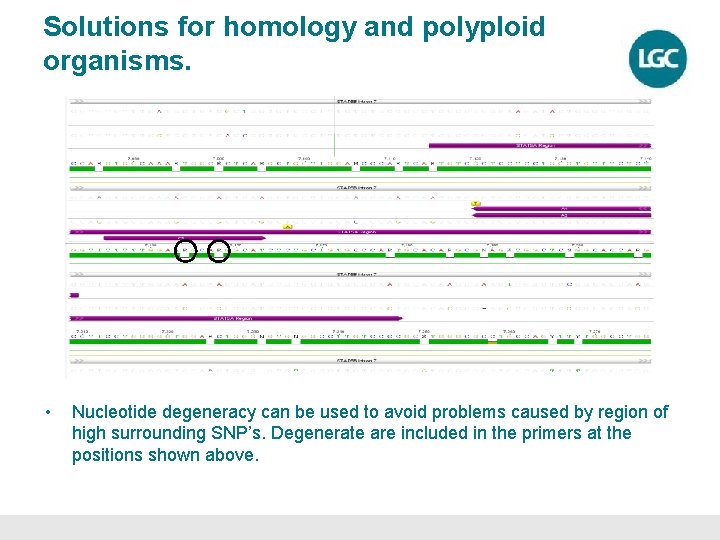
Solutions for homology and polyploid organisms. • Nucleotide degeneracy can be used to avoid problems caused by region of high surrounding SNP’s. Degenerate are included in the primers at the positions shown above.

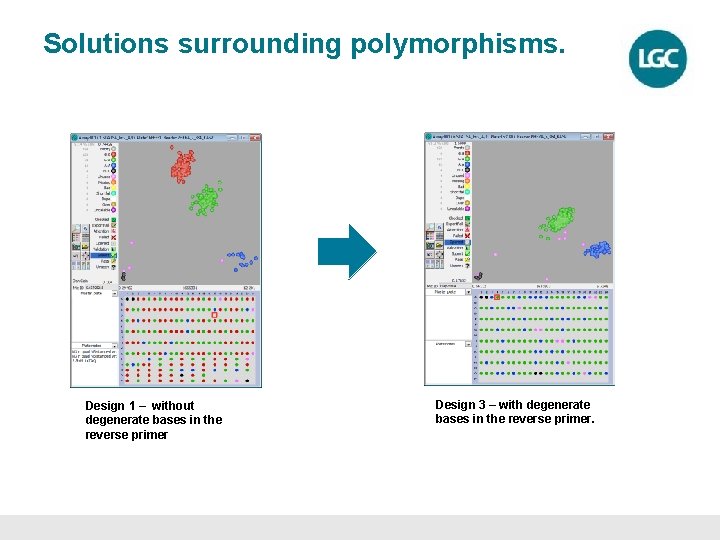
Solutions surrounding polymorphisms. Design 1 – without degenerate bases in the reverse primer Design 3 – with degenerate bases in the reverse primer.