John 1 1 3 1 2 3 In


![The Universal Language “[The universe] is written in the language of mathematics, and its The Universal Language “[The universe] is written in the language of mathematics, and its](https://slidetodoc.com/presentation_image/d1e0ac2a4bba327f2df54395f0d63e41/image-4.jpg)




















- Slides: 51

John 1: 1 -3 1 2 3 In the beginning was the Word, and the Word was with God, and the Word was God. The same was in the beginning with God. All things were made by him; and without him was not any thing made that was made. © 2001 Timothy G. Standish

The Genetic Code: The Language of Life Timothy G. Standish, Ph. D. © 2001 Timothy G. Standish

Information Only Goes One Way The central dogma states that once “information” has passed into protein it cannot get out again. The transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein, may be possible, but transfer from protein to protein, or from protein to nucleic acid, is impossible. Information means here the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein. Francis Crick, 1958 © 2001 Timothy G. Standish
![The Universal Language The universe is written in the language of mathematics and its The Universal Language “[The universe] is written in the language of mathematics, and its](https://slidetodoc.com/presentation_image/d1e0ac2a4bba327f2df54395f0d63e41/image-4.jpg)
The Universal Language “[The universe] is written in the language of mathematics, and its characters are triangles, circles, and other geometrical figures, without which it is humanly impossible to understand a single word of it. . ” Galileo Galilei, The Controversy on the Comets of 1618 © 2001 Timothy G. Standish

The Genetic Language �The genetic code is a written language not unlike English or German �While English uses 26 letters to spell out words, genetic languages use only 4 nucleotide “letters” �The DNA nucleotide language is transcribed into the RNA nucleotide language �The nucleotide language must be translated into the amino acid language to make proteins © 2001 Timothy G. Standish

The Nucleotide Language - ATGCATGC � RNA - AUGCAUGC � It is not unlike different Bible versions. � Psalm 139: 14 � KJV I will praise thee; for I am fearfully and wonderfully made: marvelous are thy works; and that my soul knoweth right well. � NIV I praise you because I am fearfully and wonderfully made; your works are wonderful, I know that full well. � DNA © 2001 Timothy G. Standish

Nucleotide Words �Words in the nucleotide language are all 3 letters or bases long �This means that there can only be 43 = 64 unique words �If each codon was only 2 bases long, there would be 42 = 16 possible unique codons �This would not provide enough unique meanings to code for the 22 things (20 amino acids plus start and stop) that have to be coded for. © 2001 Timothy G. Standish

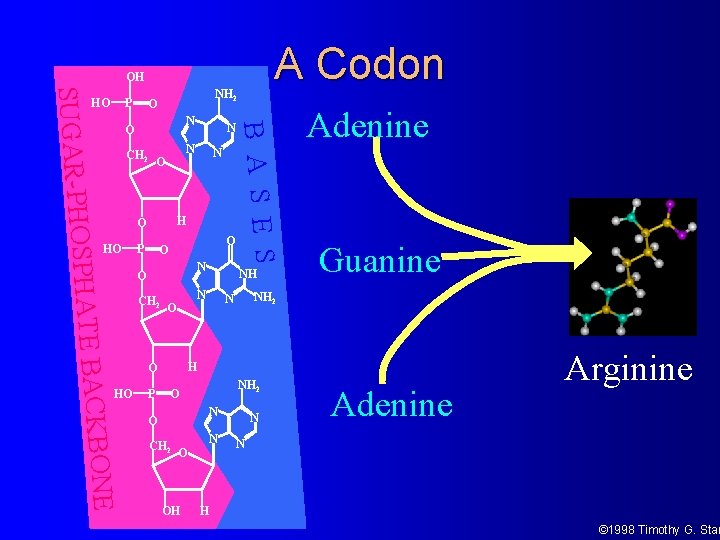
A Codon OH NH 2 O N O CH 2 N O P O O PHATE BACK N O CH 2 P NH N O Adenine Guanine NH 2 N H O HO N B A S E S SUGAR-PHOS P HO NH 2 O N O BONE CH 2 O OH N N Adenine Arginine N H © 1998 Timothy G. Stan

The Genetic Code Helps To Control The Impact Of Point Mutations © 2001 Timothy G. Standish

Redundancy in the Code �Codons code for only 20 words, or amino acids. �In addition to the amino acids, the start and stop of a protein need to be coded for �There are thus a total of 22 unique meanings for the 64 codons �Because all codons are used many act as synonyms, having different sequences, but the same meaning �The fact that many amino acids are coded for by several codons is called degeneracy © 2001 Timothy G. Standish

�Genes Sentences can be thought of as sentences in the nucleic acid language �Each gene contains a sequence of codons that describe the primary structure (amino acid sequence) of a polypeptide (protein). �At the beginning of each gene, acting like a capital letter is the start codon �In the middle is a sequence of codons for amino acids �The period at the end is a stop codon © 2001 Timothy G. Standish

�The Protein Language protein language is very different from the nucleotide language �Polypeptides are the sentences �It is analogous to pictographic languages like Chinese or Egyptian Hieroglyphics. �Each symbol has a meaning in pictographic languages; and in proteins, each amino acid has a unique meaning or specific effect. �Words are not a sequence of nucleotides, but each AA in the primary structure © 2001 Timothy G. Standish

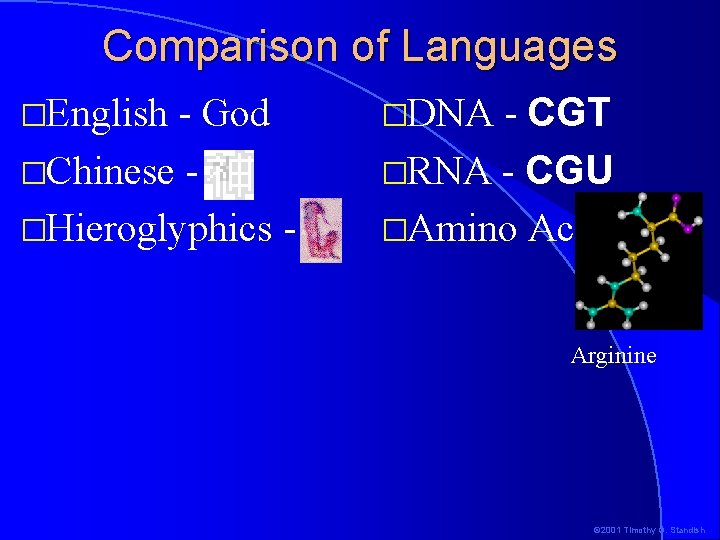
Comparison of Languages �English - God �Chinese �Hieroglyphics - - CGT �RNA - CGU �Amino Acid �DNA Arginine © 2001 Timothy G. Standish

Redundancy: Synonyms and Codon Degeneracy �English - Synonyms �Nucleic acids for God: Synonyms for Arginine: �Lord �CGU �Father �CGC �Deity �CGA �the Almighty �CGG �Jehovah �AGA �AGG © 2001 Timothy G. Standish

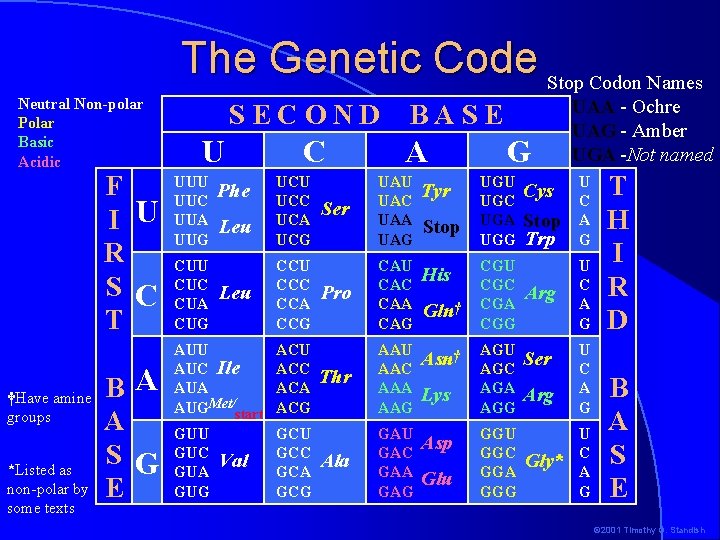
The Genetic Code Stop Codon Names Neutral Non-polar Polar Basic Acidic F I U R S C T †Have amine groups *Listed as non-polar by some texts B A A S G E SECOND U UUC UUA UUG CUU CUC CUA CUG Phe Leu C UCU UCC UCA UCG CCU CCC CCA CCG AUU AUC Ile AUA AUGMet/start ACU ACC ACA ACG GUU GUC GUA GUG GCU GCC GCA GCG Val BASE A Ser UAU UAC UAA UAG Tyr Pro CAU CAC CAA CAG His Thr AAU AAC AAA AAG Asn† Ala GAU GAC GAA GAG Asp Stop Gln† Lys Glu G UGU UGC UGA UGG CGU CGC CGA CGG AGU AGC AGA AGG GGU GGC GGA GGG UAA - Ochre UAG - Amber UGA -Not named Cys Stop Trp U C A G Arg U C A G Ser Arg Gly* U C A G T H I R D B A S E © 2001 Timothy G. Standish

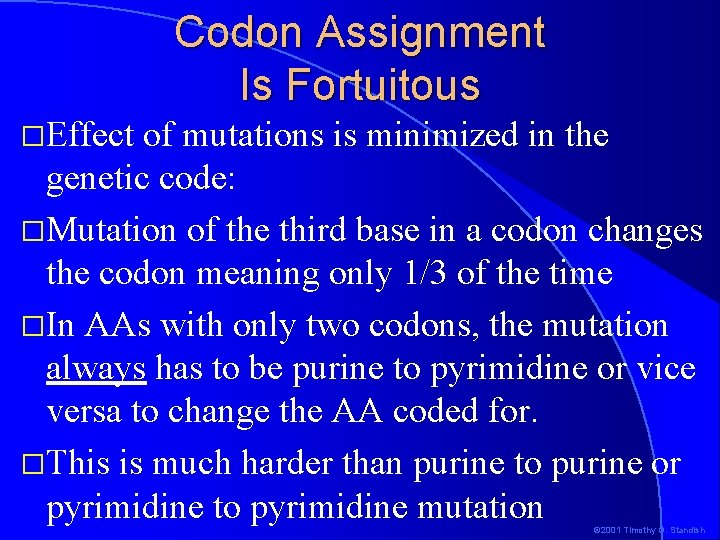
Codon Assignment Is Fortuitous �Effect of mutations is minimized in the genetic code: �Mutation of the third base in a codon changes the codon meaning only 1/3 of the time �In AAs with only two codons, the mutation always has to be purine to pyrimidine or vice versa to change the AA coded for. �This is much harder than purine to purine or pyrimidine to pyrimidine mutation © 2001 Timothy G. Standish

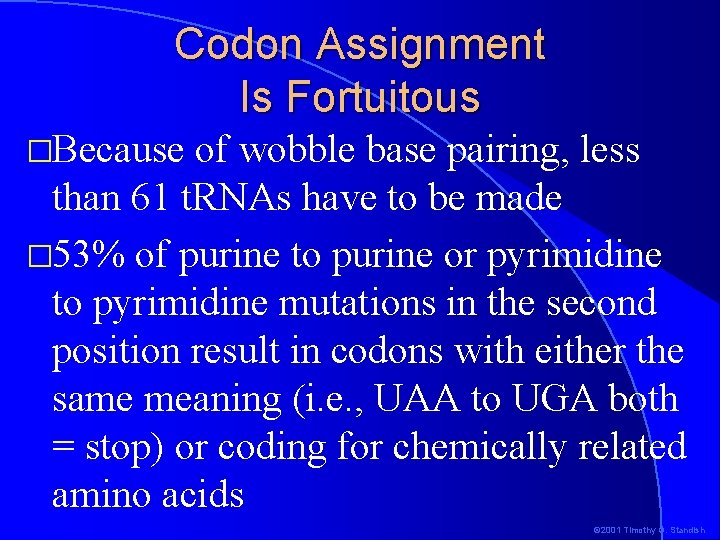
Codon Assignment Is Fortuitous �Because of wobble base pairing, less than 61 t. RNAs have to be made � 53% of purine to purine or pyrimidine to pyrimidine mutations in the second position result in codons with either the same meaning (i. e. , UAA to UGA both = stop) or coding for chemically related amino acids © 2001 Timothy G. Standish

The Genetic Code Is Improbable And Does Not Look Random © 2001 Timothy G. Standish

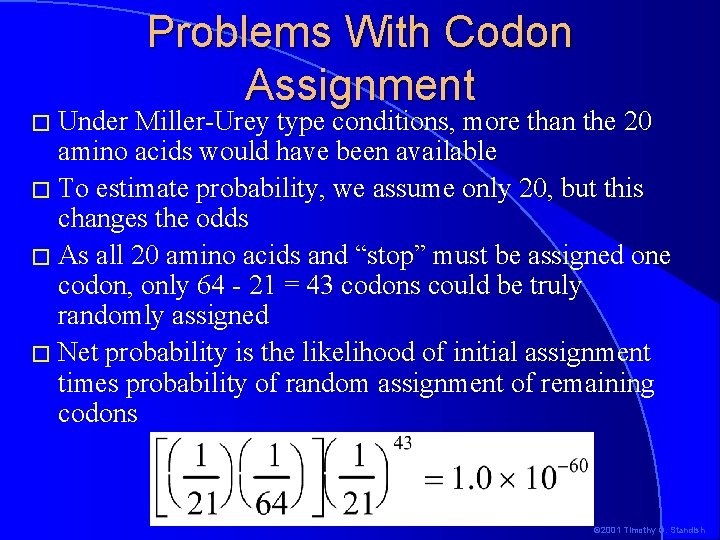
Possible Codon Assignments � The probability of getting the assignment of codons to amino acids we have can be calculated as follows: – There are 21 meanings for codons: 20 amino acids � 1 stop � 1 start, which doesn’t count because it also is assigned to methionine � – 64 Codons � If we say that each codon has an equal probability of being assigned to an amino acid, then the probability of getting any particular set of 64 assignments is: or 0. 0000000000000000000000000 0000024 © 2001 Timothy G. Standish

� Under Problems With Codon Assignment Miller-Urey type conditions, more than the 20 amino acids would have been available � To estimate probability, we assume only 20, but this changes the odds � As all 20 amino acids and “stop” must be assigned one codon, only 64 - 21 = 43 codons could be truly randomly assigned � Net probability is the likelihood of initial assignment times probability of random assignment of remaining codons © 2001 Timothy G. Standish

Initial Codon Assignment � Theory would indicate initial codon assignment must have been random � Lewin in Genes VI, p 214, 215 suggests the following scenario: 1 A small number of codons randomly get meanings representing a few amino acids or possibly one codon representing a “group” of amino acids 2 More precise codon meaning evolves perhaps with only the first two bases having meaning with discrimination at the third position evolving later 3 The code becomes “frozen” when the system becomes so complex that changes in codon meaning would disrupt existing vital proteins © 2001 Timothy G. Standish

Codon Assignment does not look random The genetic code does not like uneven numbers. © 2001 Timothy G. Standish

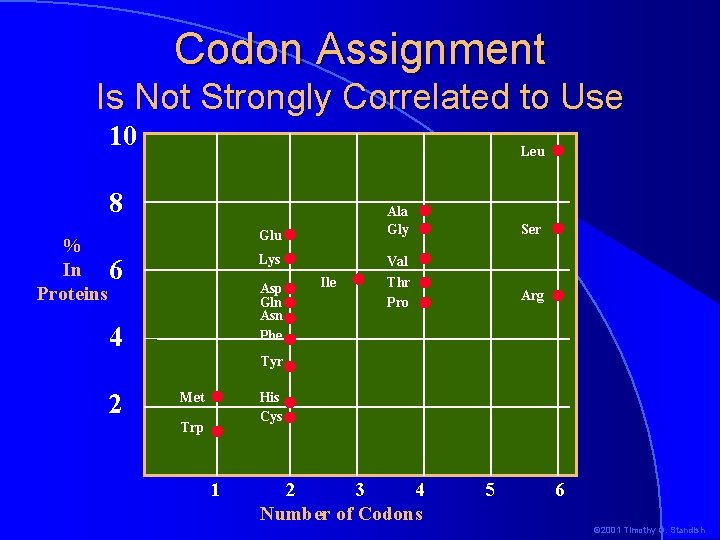
�If Initial Codon Assignment natural selection worked on codons, the most commonly used amino acids might be expected to have the most codons �If there was some sort of random assignment, the same thing might be expected �This is not the case © 2001 Timothy G. Standish

Codon Assignment Is Not Strongly Correlated to Use 10 Leu 8 Glu % In 6 Proteins Lys Asp Gln Asn Phe 4 Ile Ala Gly Ser Val Thr Pro Arg Tyr 2 Met His Cys Trp 1 2 3 4 Number of Codons 5 6 © 2001 Timothy G. Standish

The Genetic Code Is Not Completely Universal © 2001 Timothy G. Standish

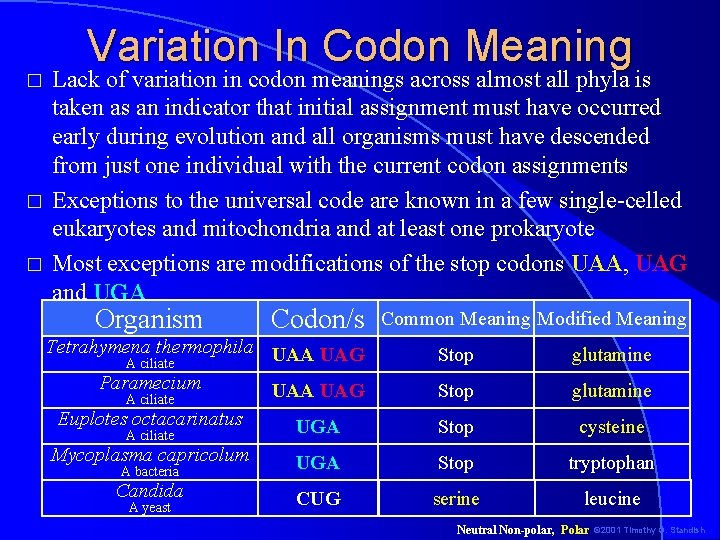
� � � Variation In Codon Meaning Lack of variation in codon meanings across almost all phyla is taken as an indicator that initial assignment must have occurred early during evolution and all organisms must have descended from just one individual with the current codon assignments Exceptions to the universal code are known in a few single-celled eukaryotes and mitochondria and at least one prokaryote Most exceptions are modifications of the stop codons UAA, UAG and UGA Organism Codon/s Tetrahymena thermophila UAA UAG A ciliate Paramecium UAA UAG A ciliate Common Meaning Modified Meaning Stop glutamine Euplotes octacarinatus UGA Stop cysteine Mycoplasma capricolum UGA Stop tryptophan Candida CUG serine leucine A ciliate A bacteria A yeast Neutral Non-polar, Polar © 2001 Timothy G. Standish

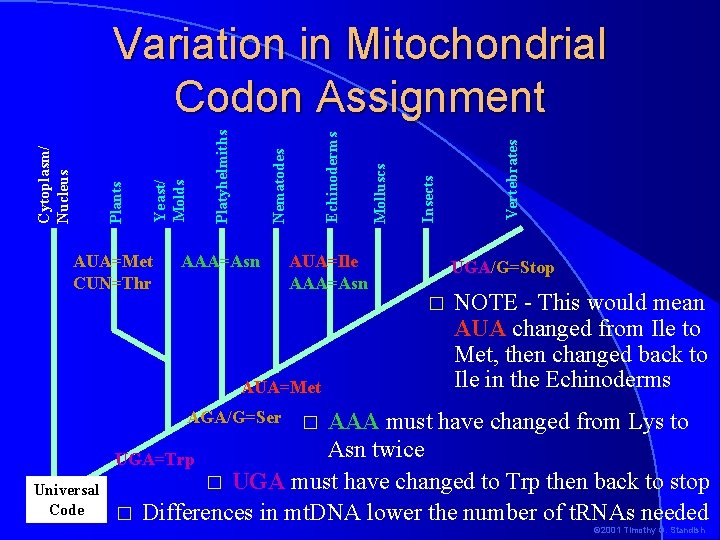
AUA=Met CUN=Thr AAA=Asn AUA=Ile AAA=Asn UGA/G=Stop � AUA=Met NOTE - This would mean AUA changed from Ile to Met, then changed back to Ile in the Echinoderms AAA must have changed from Lys to Asn twice UGA=Trp � UGA must have changed to Trp then back to stop � Differences in mt. DNA lower the number of t. RNAs needed AGA/G=Ser Universal Code Vertebrates Insects Molluscs Echinoderms Nematodes Platyhelmiths Yeast/ Molds Plants Cytoplasm/ Nucleus Variation in Mitochondrial Codon Assignment � © 2001 Timothy G. Standish

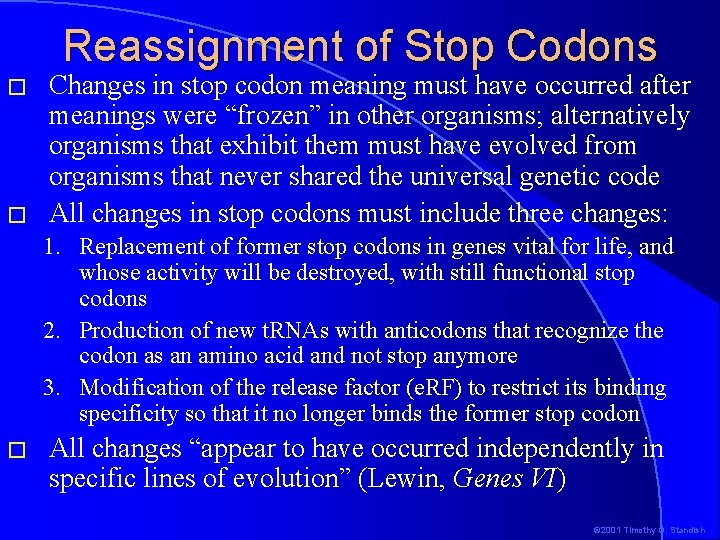
Reassignment of Stop Codons � � Changes in stop codon meaning must have occurred after meanings were “frozen” in other organisms; alternatively organisms that exhibit them must have evolved from organisms that never shared the universal genetic code All changes in stop codons must include three changes: 1. Replacement of former stop codons in genes vital for life, and whose activity will be destroyed, with still functional stop codons 2. Production of new t. RNAs with anticodons that recognize the codon as an amino acid and not stop anymore 3. Modification of the release factor (e. RF) to restrict its binding specificity so that it no longer binds the former stop codon � All changes “appear to have occurred independently in specific lines of evolution” (Lewin, Genes VI) © 2001 Timothy G. Standish

� Once Changing Initial Codon Assignment codons have been assigned to an amino acid, changing their meaning would require: – Changing the t. RNA anticodon or, much harder, changing the aminoacyl-t. RNA synthetase – Changing all codons to be reassigned in at least the vital positions in those proteins needed for survival � This seems unlikely � The situation is complicated in cases where genes seem to have been swapped between the nucleus and mitochondria © 2001 Timothy G. Standish

Wobble Base Pairing © 2001 Timothy G. Standish

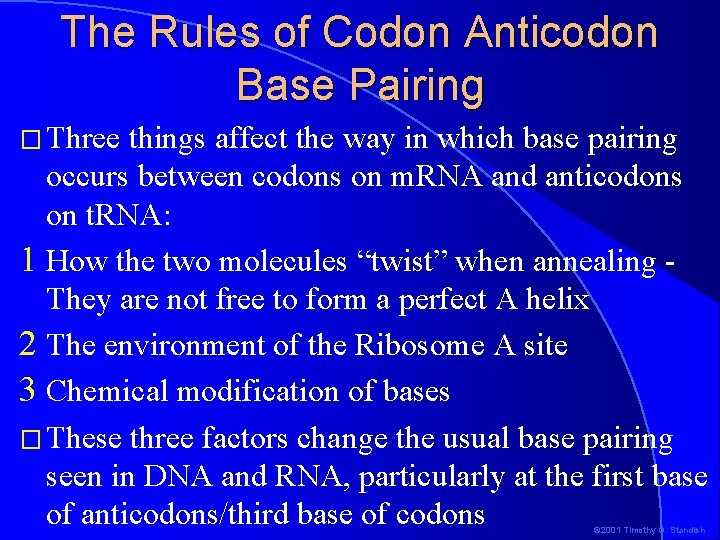
The Rules of Codon Anticodon Base Pairing � Three things affect the way in which base pairing occurs between codons on m. RNA and anticodons on t. RNA: 1 How the two molecules “twist” when annealing They are not free to form a perfect A helix 2 The environment of the Ribosome A site 3 Chemical modification of bases � These three factors change the usual base pairing seen in DNA and RNA, particularly at the first base of anticodons/third base of codons © 2001 Timothy G. Standish

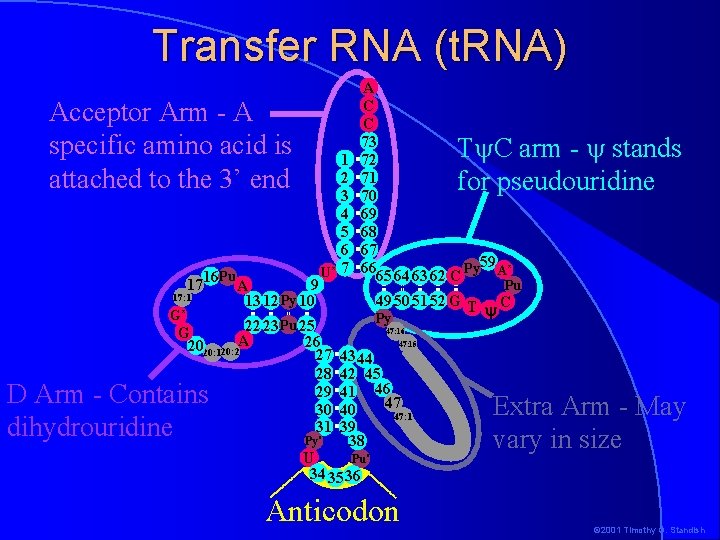
Transfer RNA (t. RNA) Acceptor Arm - A specific amino acid is attached to the 3’ end Pu 1716 A 9 17: 1 13 12 Py 10 1 2 3 4 5 6 U* 7 A C C 73 72 71 70 69 68 67 Py 59 A* 66 65 64 63 62 C D Arm - Contains dihydrouridine Py* Pu 49 50 51 52 G T C y Py G* 22 23 Pu 25 G 26 2020: 120: 2 A 27 1 28 29 30 31 T C arm - stands for pseudouridine 47: 16 47: 15 43 44 42 45 41 46 47 40 47: 1 39 38 Pu* U 34 35 36 Anticodon Extra Arm - May vary in size © 2001 Timothy G. Standish

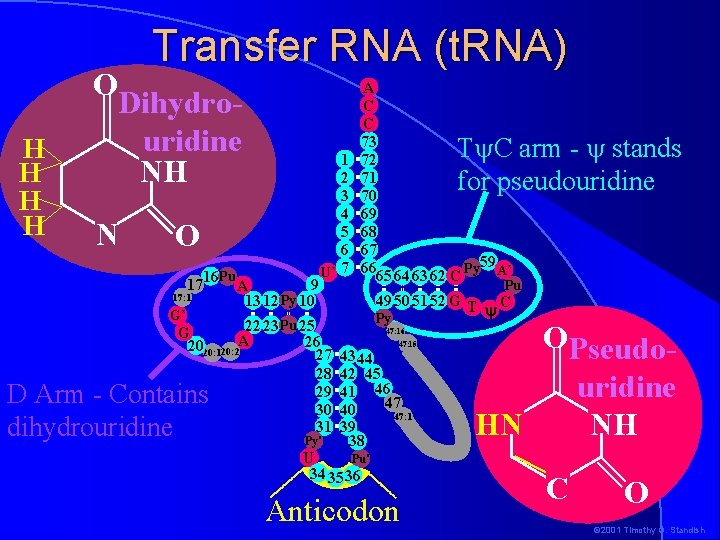
Transfer RNA (t. RNA) H H O Dihydrouridine NH N O Pu 1716 A 9 17: 1 13 12 Py 10 1 2 3 4 5 6 U* 7 A C C 73 72 71 70 69 68 67 Py 59 A* 66 65 64 63 62 C D Arm - Contains dihydrouridine Py* Pu 49 50 51 52 G T C y Py G* 22 23 Pu 25 G 26 2020: 120: 2 A 27 1 28 29 30 31 T C arm - stands for pseudouridine 47: 16 47: 15 43 44 42 45 41 46 47 40 47: 1 39 38 Pu* U 34 35 36 Anticodon OPseudouridine NH HN C O © 2001 Timothy G. Standish

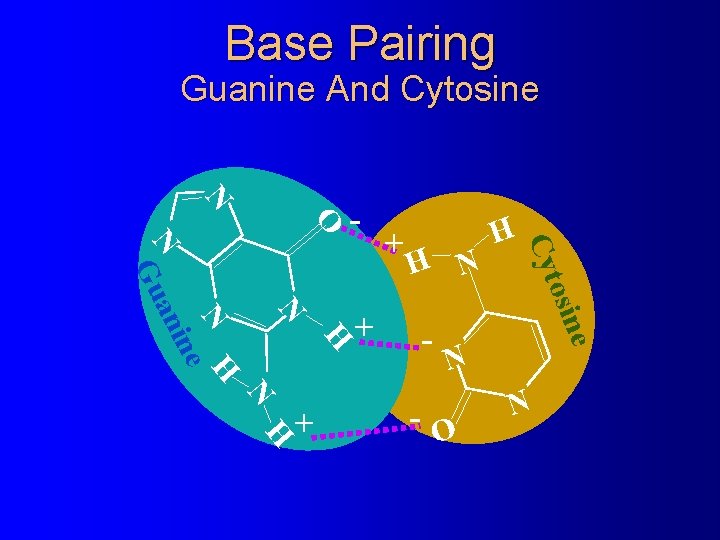
Base Pairing Guanine And Cytosine N O - H N H e osin N N H ne ani - H Cyt N Gu N H + + + N -O N

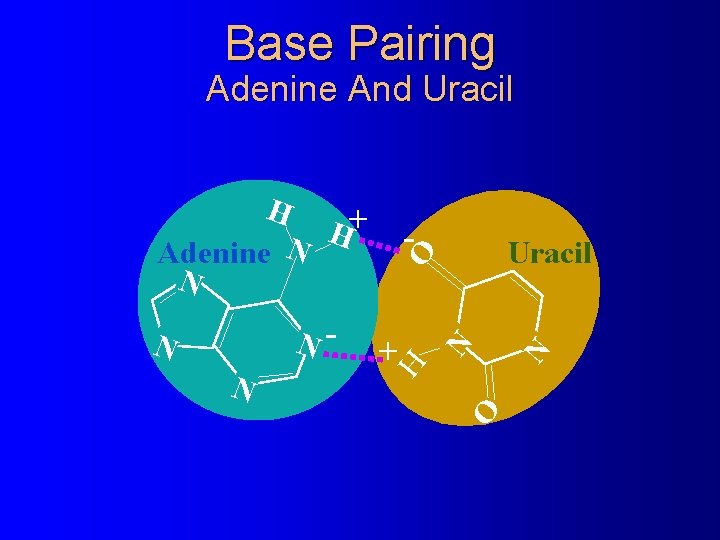
Base Pairing Adenine And Uracil N O + O N Uracil N N- N - H H + H Adenine N N

Base Pairing Adenine And Cytosine e osin Cyt H H H+ + H N N e n i n e d A N N -O N N

Base Pairing Guanine And Uracil O - N Uracil O N - N N N O H + H N N H ne ani Gu H + +

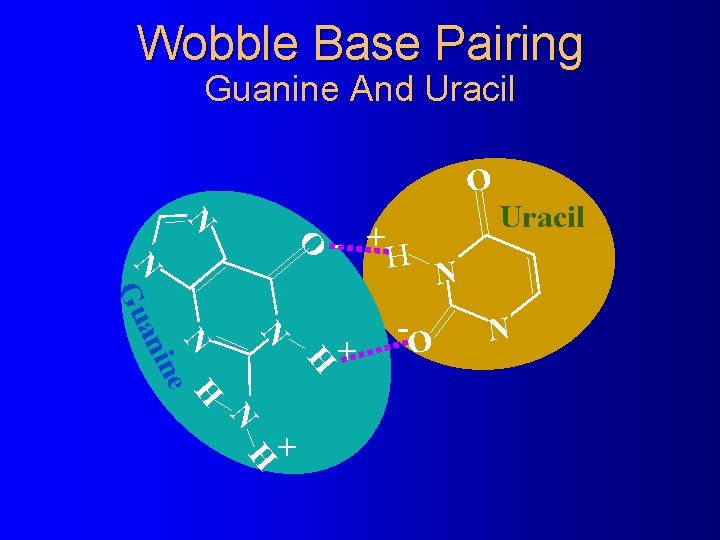
Wobble Base Pairing Guanine And Uracil O N N - +H N -O N + H N H ne ani Gu N H + Uracil

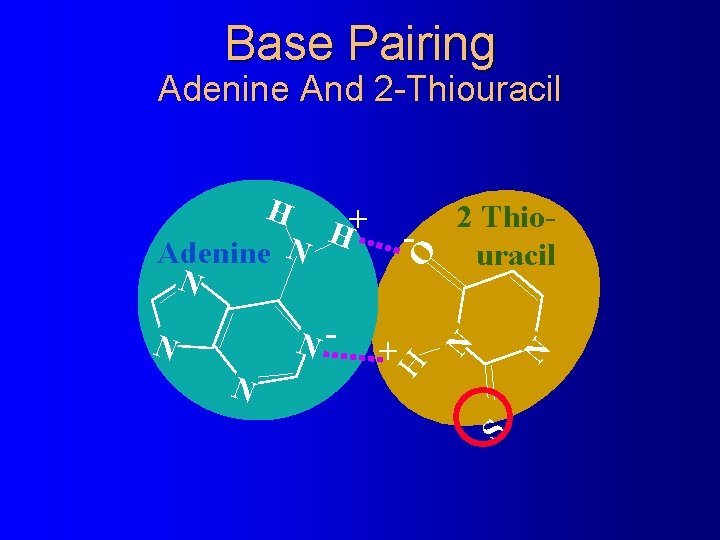
Base Pairing Adenine And 2 -Thiouracil S N N O + N N- N - 2 Thiouracil H H + H Adenine N N

Wobble Base Pairing Guanine And 2 -Thiouracil N O N - + H N H ne ani Gu N H + + S N O 2 Thiouracil N 2 -Thiouracil forms only one hydrogen bond with guanine which is not enough to form a stable pair in the environment of the ribosome A site

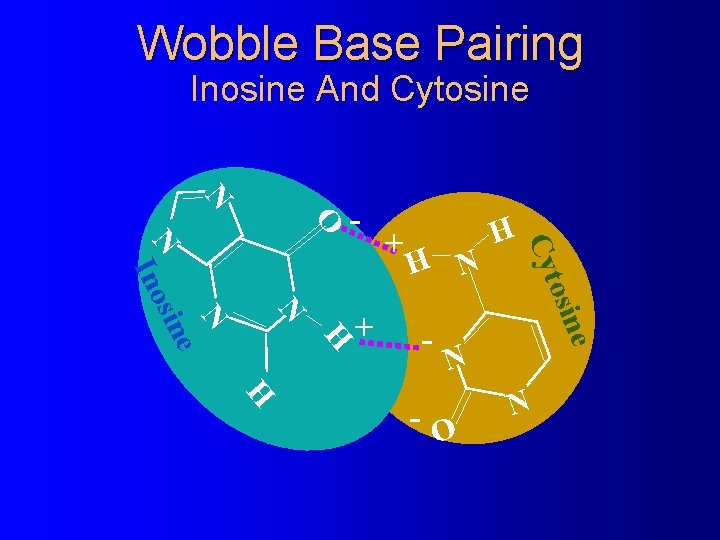
Wobble Base Pairing Inosine And Cytosine N O - H N H e osin N N e sin - H Cyt N Ino + + N H -O N

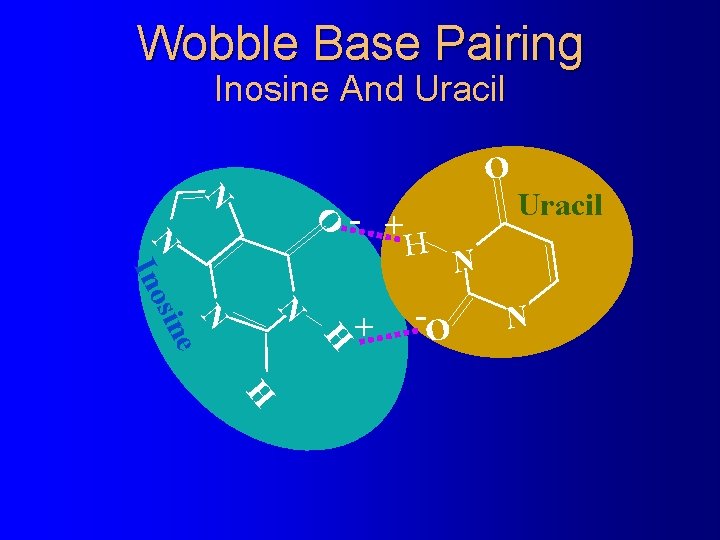
Wobble Base Pairing Inosine And Uracil O N - + N H N e sin Ino + H -O Uracil N N H

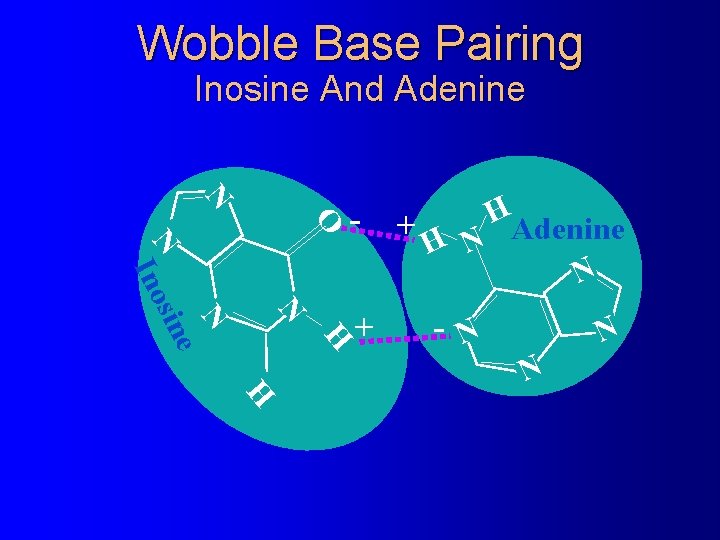
Wobble Base Pairing Inosine And Adenine N O - N N H N e sin Ino + H + N Adenine H N - N N N H

The Wacky Rules of Wobble Base Pairing Third codon base: First anticodon base: �U -------A or G -------A �C -------G �A -------U �G -------C or U �I -------C U or G � 2 -S-U © 2001 Timothy G. Standish

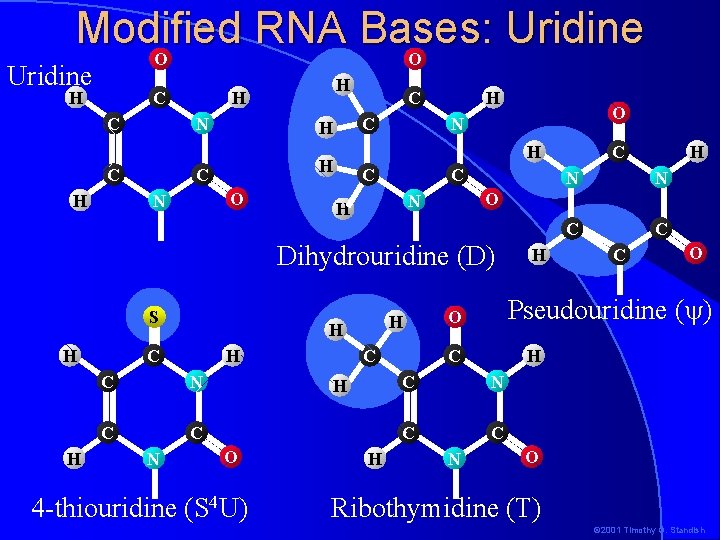
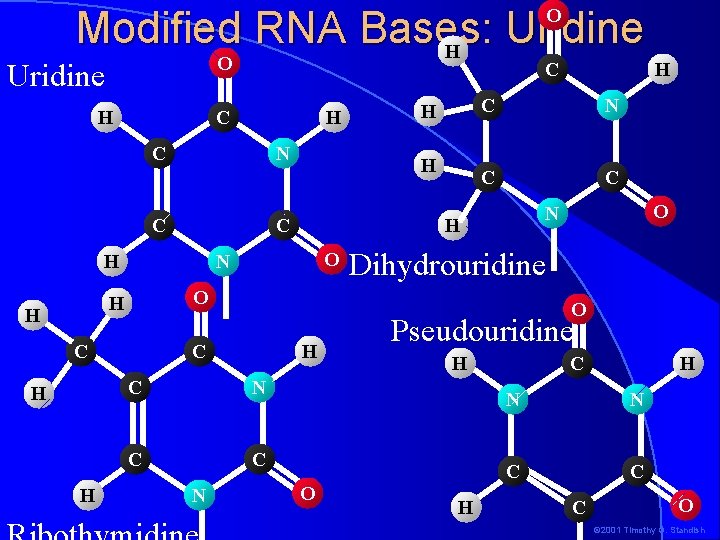
Modified RNA Bases: Uridine O O Uridine H C C N C H H C C H H H C N C C N H C H O 4 -thiouridine (S 4 U) H C H N N C C O N H Dihydrouridine (D) S O N H H O N C H C C C O Pseudouridine ( ) O C H C N C C N H O Ribothymidine (T) © 2001 Timothy G. Standish

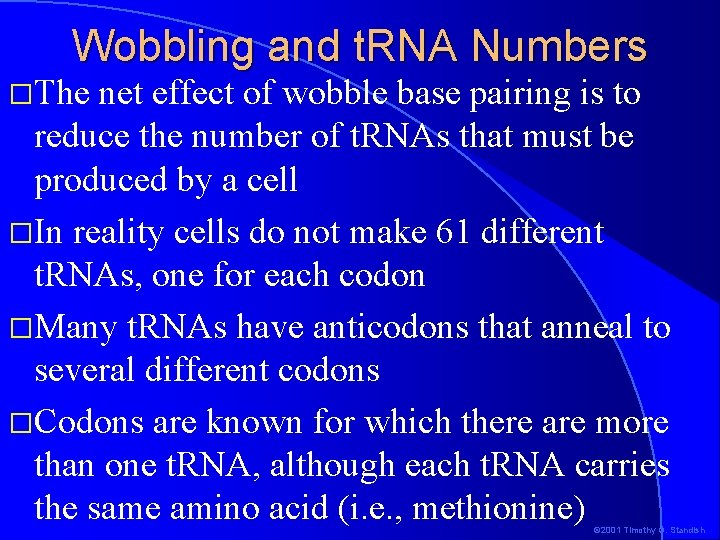
Wobbling and t. RNA Numbers �The net effect of wobble base pairing is to reduce the number of t. RNAs that must be produced by a cell �In reality cells do not make 61 different t. RNAs, one for each codon �Many t. RNAs have anticodons that anneal to several different codons �Codons are known for which there are more than one t. RNA, although each t. RNA carries the same amino acid (i. e. , methionine) © 2001 Timothy G. Standish

Summary: Are Codons The Language of God? � The genetic code appears to be non-random in nature and designed with considerable safeguards against harmful point mutations � An evolutionary model suggests at least at some level of randomness in assignment of amino acids to codons � No mechanism exists for genetic code evolution � Thus variation in the genetic code suggests a polyphyletic origin for life � Taken together, this evidence indicates the hand of a Designer in the genetic code and does not support theory that life originated due to random processes or that all organisms share a common ancestor © 2001 Timothy G. Standish

© 2001 Timothy G. Standish

Modified RNA Bases: Uridine H O O Uridine H C C H C N C H N C C O Dihydrouridine O C H C N C C N O C H N H H H C H H H O Pseudouridine H H C H N N C C C O © 2001 Timothy G. Standish

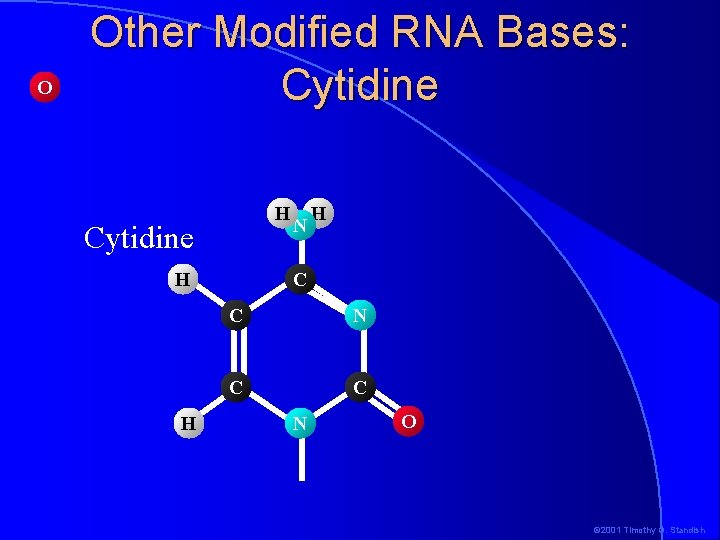
O Other Modified RNA Bases: Cytidine H H N H C C N O © 2001 Timothy G. Standish

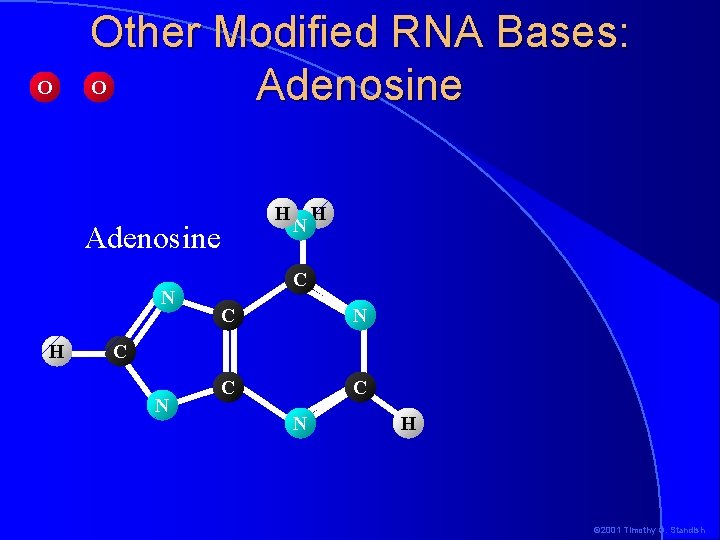
O Other Modified RNA Bases: O Adenosine H Adenosine N H C C N C C C N N H © 2001 Timothy G. Standish