IV Variation in Quantitative Traits A Quantitative Effects




































- Slides: 57

IV. Variation in Quantitative Traits A. Quantitative Effects

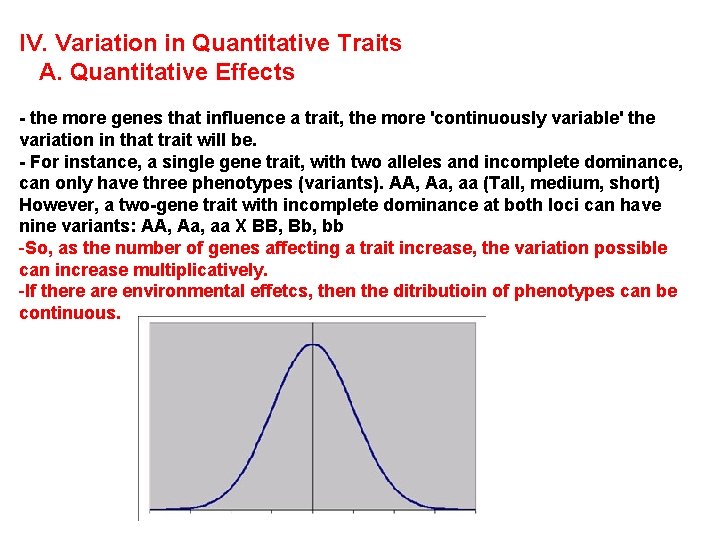
IV. Variation in Quantitative Traits A. Quantitative Effects - the more factors that influence a trait (genetic and environmental) , the more 'continuously variable' the variation in that trait will be.

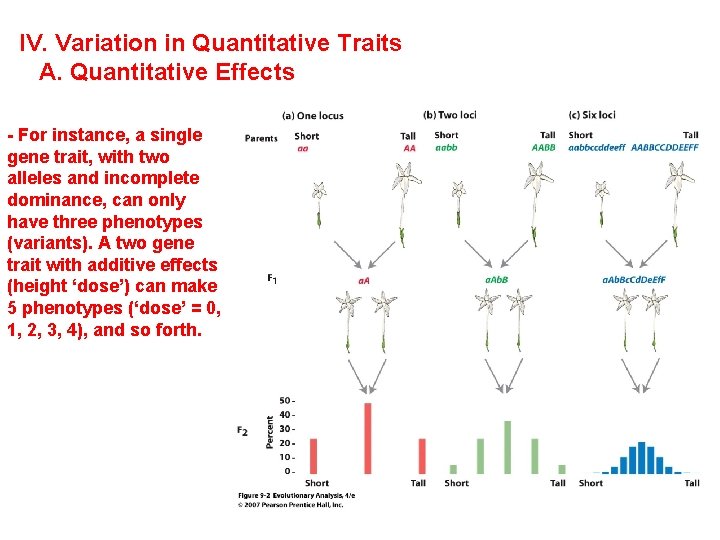
IV. Variation in Quantitative Traits A. Quantitative Effects - For instance, a single gene trait, with two alleles and incomplete dominance, can only have three phenotypes (variants). A two gene trait with additive effects (height ‘dose’) can make 5 phenotypes (‘dose’ = 0, 1, 2, 3, 4), and so forth.

IV. Variation in Quantitative Traits A. Quantitative Effects - the more genes that influence a trait, the more 'continuously variable' the variation in that trait will be. - For instance, a single gene trait, with two alleles and incomplete dominance, can only have three phenotypes (variants). AA, Aa, aa (Tall, medium, short) However, a two-gene trait with incomplete dominance at both loci can have nine variants: AA, Aa, aa X BB, Bb, bb - So, as the number of genes affecting a trait increase, the variation possible can increase multiplicatively.

IV. Variation in Quantitative Traits A. Quantitative Effects - the more genes that influence a trait, the more 'continuously variable' the variation in that trait will be. - For instance, a single gene trait, with two alleles and incomplete dominance, can only have three phenotypes (variants). AA, Aa, aa (Tall, medium, short) However, a two-gene trait with incomplete dominance at both loci can have nine variants: AA, Aa, aa X BB, Bb, bb -So, as the number of genes affecting a trait increase, the variation possible can increase multiplicatively. -If there are environmental effetcs, then the ditributioin of phenotypes can be continuous.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance - The phenotypic variation that we see in continuous traits is due to a number of factors that can be "lumped" as environmental or genetic. V(phen) = V(env) + V(gen)

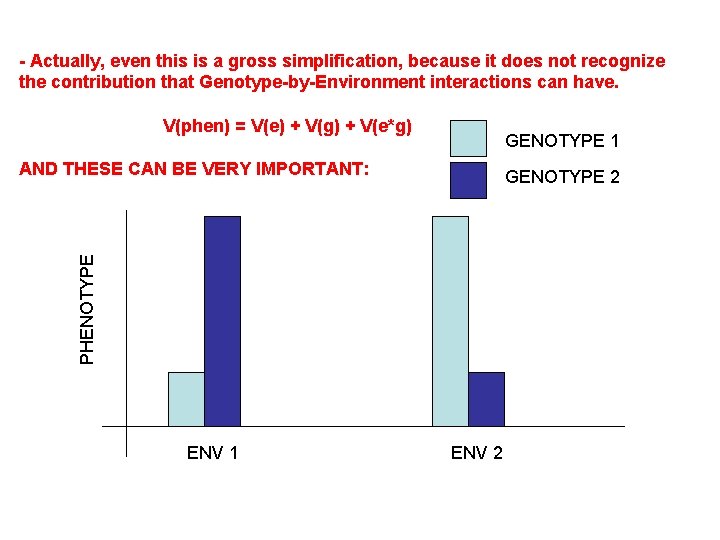
IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance - The phenotypic variation that we see in continuous traits is due to a number of factors that can be "lumped" as environmental or genetic. V(phen) = V(env) + V(gen) - Actually, even this is a gross simplification, because it does not recognize the contribution that Genotype-by-Environment interactions can have. V(phen) = V(e) + V(g) + V(e*g)

- Actually, even this is a gross simplification, because it does not recognize the contribution that Genotype-by-Environment interactions can have. V(phen) = V(e) + V(g) + V(e*g) GENOTYPE 1 AND THESE CAN BE VERY IMPORTANT: PHENOTYPE GENOTYPE 2 ENV 1 ENV 2

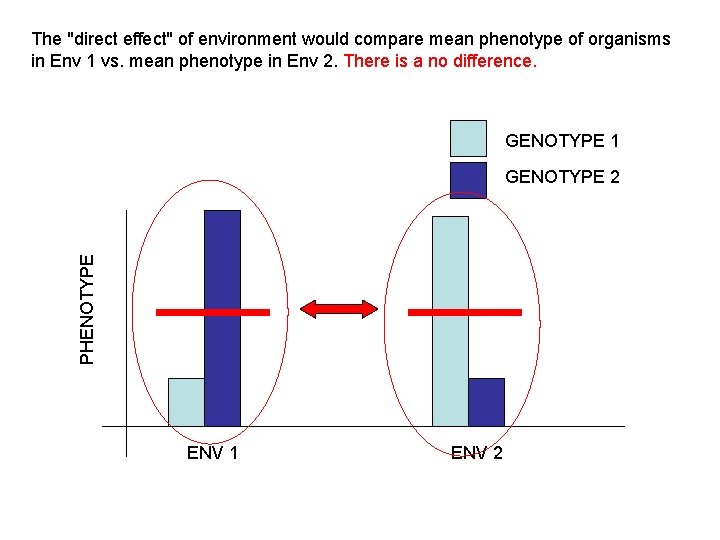
The "direct effect" of environment would compare mean phenotype of organisms in Env 1 vs. mean phenotype in Env 2. There is a no difference. GENOTYPE 1 PHENOTYPE GENOTYPE 2 ENV 1 ENV 2

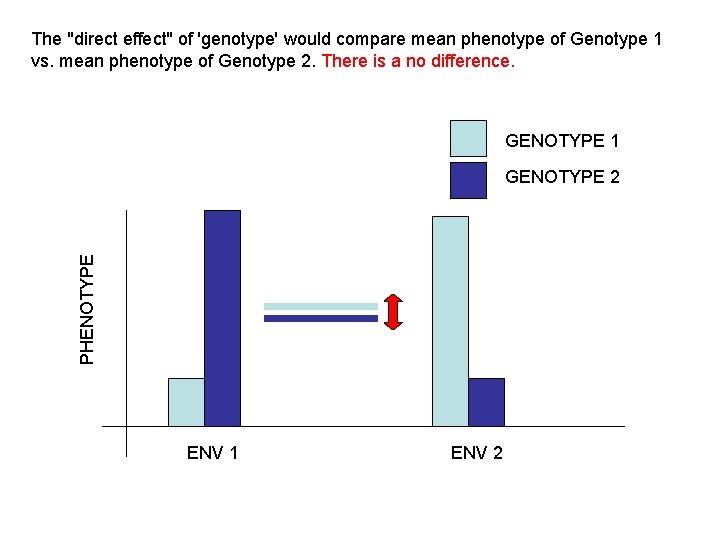
The "direct effect" of 'genotype' would compare mean phenotype of Genotype 1 vs. mean phenotype of Genotype 2. There is a no difference. GENOTYPE 1 PHENOTYPE GENOTYPE 2 ENV 1 ENV 2

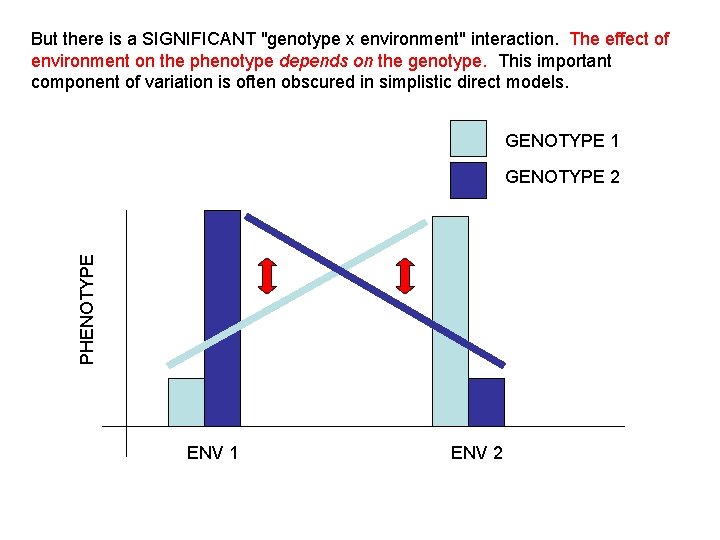
But there is a SIGNIFICANT "genotype x environment" interaction. The effect of environment on the phenotype depends on the genotype. This important component of variation is often obscured in simplistic direct models. GENOTYPE 1 PHENOTYPE GENOTYPE 2 ENV 1 ENV 2

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance - The phenotypic variation that we see in continuous traits is due to a number of factors that can be "lumped" as environmental or genetic. V(phen) = V(env) + V(gen) - Actually, even this is a gross simplification, because it does not recognize the contribution that Genotype-by-Environment interactions can have. V(phen) = V(e) + V(g) + V(e*g) - Ultimately, the goal of evolutionary studies is to determine the contribution of genetic variation, because this is the only variation that is heritable and can evolve. “Broad-sense” heritability = Vg/Vp

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation - Even the genetic variation is more complex than one might think. There is variation due to 'additive' genetic variance, 'dominance' genetic variance, 'epistasis', and a variety of other contributors (sex linkage) that can be modeled. V(g) = V(a) + V(d) + V(ep)

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation - Even the genetic variation is more complex than one might think. There is variation due to 'additive' genetic variance, 'dominance' genetic variance, 'epistasis', and a variety of other contributors that can be modeled. - We will concern ourselves with 'additive variation' Think of an individual that is AA. If the 'A' allele is adaptive, then their fitness will be higher than the mean fitness of the population. Their offspring, as a consequence of inheriting this adaptive gene, will also have a higher fitness than the population, as a whole. This allele 'adds' fitness. 2 A’s (AA) adds more…

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Broad-sense (H) = V(g)/V(p) - difficult to measure

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Broad-sense (H) = V(g)/V(p) - difficult to measure - Narrow-sense (h 2) = V(a)/V(p)

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Broad-sense (H) = V(g)/V(p) - difficult to measure - Narrow-sense (h 2) = V(a)/V(p) – easier to measure

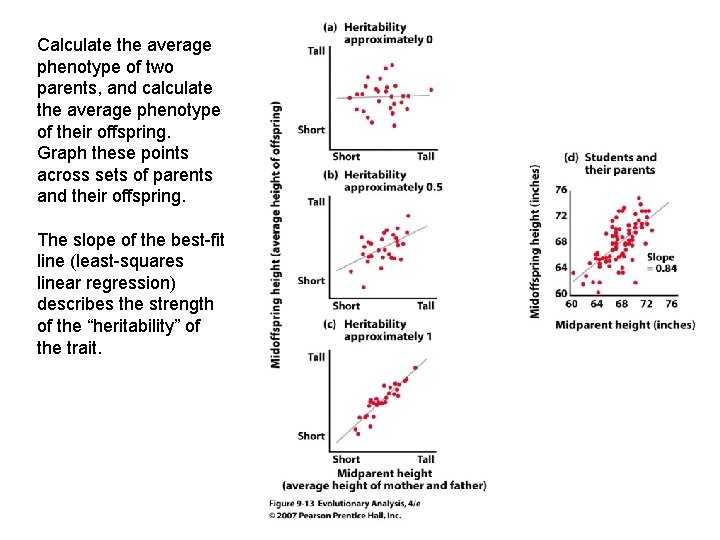
Calculate the average phenotype of two parents, and calculate the average phenotype of their offspring. Graph these points across sets of parents and their offspring. The slope of the best-fit line (least-squares linear regression) describes the strength of the “heritability” of the trait.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Calculating Heritability from Selection Experiments - Consider a variable population, with mean phenotype = x.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4)

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4 - The heritability (narrow sense) = R/S = 4/5 = 0. 8. because R = h 2 s

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4 - The heritability (narrow sense) = R/S = 4/5 = 0. 8. - The closer the offspring are to their particular parents (in amount of deviation from the whole population), the greater the heritability and the more rapid the response to selection.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - Consider a variable population, with mean phenotype = x. - Select organisms with a more extreme phenotype (x + 5) to breed. - The selection differential, S = (mean of breeding pop) - (mean of entire pop) S = (x + 5) - (x) = 5 - Suppose the offspring mean phenotype = (x + 4) - The Response to Selection = R = difference between the whole original population and the offspring: R = (X + 4) - (x) = 4 - The heritability (narrow sense) = R/S = 4/5 = 0. 8. - The closer the offspring get to their particular parents (in amount of deviation from the whole population), the greater the heritability and the more rapid the response to selection. - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember?

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember)? - So, through a series of selection experiments, we can determine how responsive a trait is to selective pressure.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember)? - So, through a series of selection experiments, we can determine how responsive a trait is to selective pressure. As selection proceeds, most variation is environmental or dominance and response to selection slows.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability - This quantifies the evolutionarily important genetic variance (heritability is also V(add)/V(phen), remember)? - So, through a series of selection experiments, we can determine how responsive a trait is to selective pressure. As selection proceeds, most variation is environmental or dominance and response to selection slows. - So, counterintuitively, adaptive traits may show low heritability. . . they have already been selected for, and most of the phneotypic variation NOW is probably environmental. EXAMPLE: Polar bears all have genetically determined white fur - it has been adaptive and has become fixed in their population. But they still vary in coat color (phenotype) as a result of dirt, etc. But the offspring of dirty bears will be just as white as the offspring of clean bears. . . no response to selection for 'dirty bears' because all the variation is environmental at this point.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability 4. Misuses of Heritability: Heritability is a property of a trait, in a given population, in a given environment. It provides no insight for comparisons across populations in different environments…. Why?

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability 4. Misuses of Heritability: Heritability is a property of a trait, in a given population, in a given environment. It provides no insight for comparisons across populations in different environments…. Why? Genotype x environment interactions…

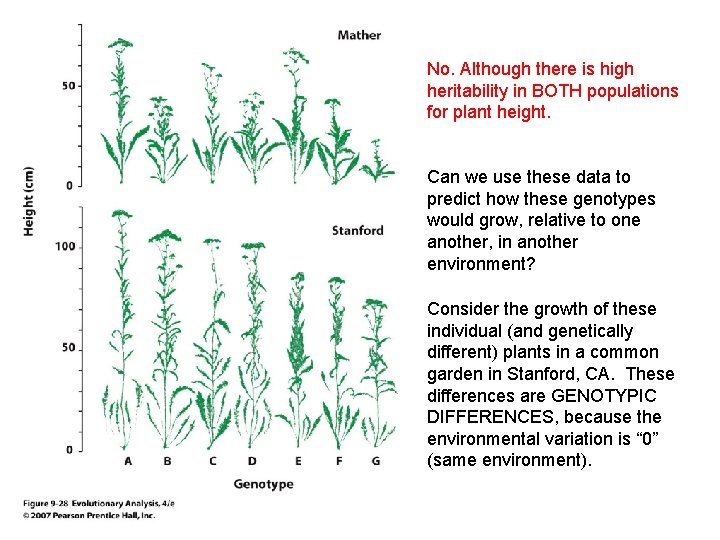
Consider the growth of these individual (and genetically different) plants in a common garden in Stanford, CA. These differences are GENOTYPIC DIFFERENCES, because the environmental variation is “ 0” (same environment).

Can we use these data to predict how these genotypes would grow, relative to one another, in another environment? Consider the growth of these individual (and genetically different) plants in a common garden in Stanford, CA. These differences are GENOTYPIC DIFFERENCES, because the environmental variation is “ 0” (same environment).

No. Although there is high heritability in BOTH populations for plant height. Can we use these data to predict how these genotypes would grow, relative to one another, in another environment? Consider the growth of these individual (and genetically different) plants in a common garden in Stanford, CA. These differences are GENOTYPIC DIFFERENCES, because the environmental variation is “ 0” (same environment).

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability 4. Misuses of Heritability: Heritability DOES NOT equal “genetically based” Many traits that are determined genetically are fixed, with no genetic variation, and so have very low heritability.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance 1. Partitioning Phenotypic Variance 2. Partitioning Genetic Variation 3. Heritability 4. Misuses of Heritability: Heritability DOES NOT equal “genetically based” Many traits that are determined genetically are fixed, with no genetic variation, and so have very low heritability. Finally, a take on twin studies: Some social psychologists beileve that we can determine “heritability” or “genetic contribution” to a triat by examining the degree of similarity between ‘monozygotic’ (identical) and ‘dizygotic’ (fraternal) twins.

Finally, a take on twin studies: Some social psychologists beileve that we can determine “heritability” or “genetic contribution” to a triat by examining the degree of similarity between ‘monozygotic’ (identical) and ‘dizygotic’ (fraternal) twins. If twins are reared apart (so that the environments are presumed to be randomized and ‘equal’ across the populations), then traits that show greater correlations between mz twins suggest a greater degree of genetic involvement.

Finally, a take on twin studies: Some social psychologists beileve that we can determine “heritability” or “genetic contribution” to a triat by examining the degree of similarity between ‘monozygotic’ (identical) and ‘dizygotic’ (fraternal) twins. If twins are reared apart (so that the environments are presumed to be randomized and ‘equal’ across the populations), then traits that show greater correlations between mz twins suggest a greater degree of genetic involvement. Problem: You don’t know that the environments are similar or different. The Jim twins: http: //science. howstuffworks. com/genetic-science/twin 1. htm

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance C. Selection on Quantitative Traits - Traits affected by many genes have a higher probability of including a pleiotrophic gene – a gene that affects more than one trait. So, we might expect complex, quantitative traits to be CORRELATED to other traits. If selection is acting on both traits in different ways, neither will be “optimized”. Adaptations will be a compromise, depending on the relative strengths of the selective pressures, the relative values of the adaptive traits, and their heritabilities (ease with which they can respond to selection).

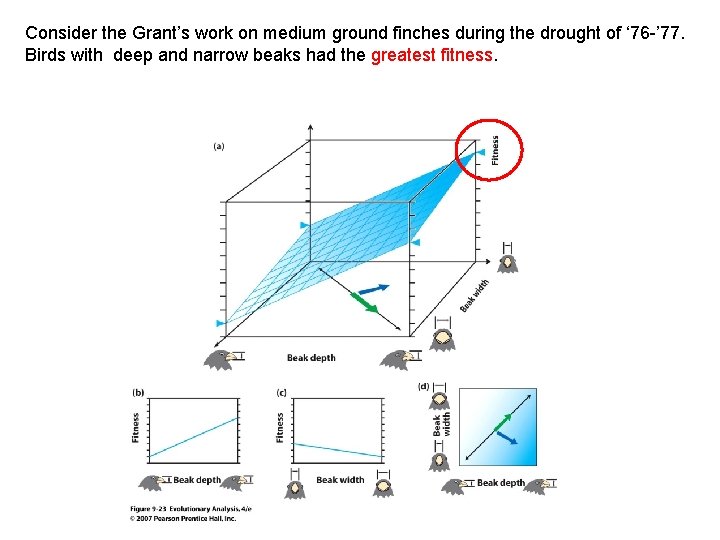
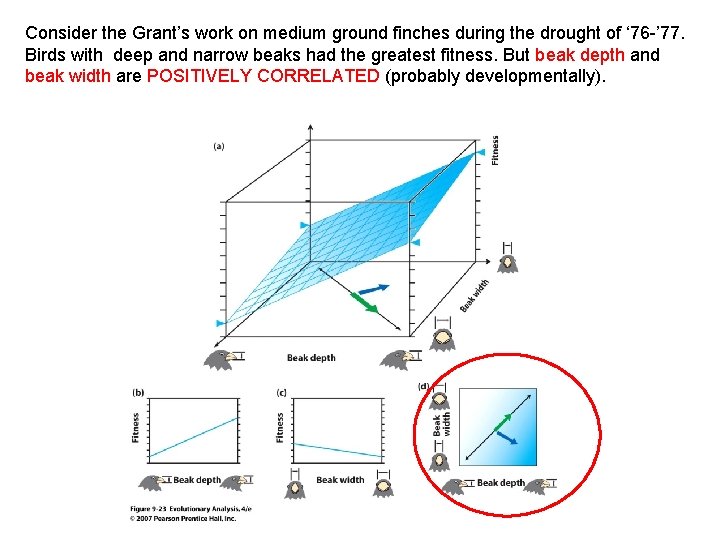
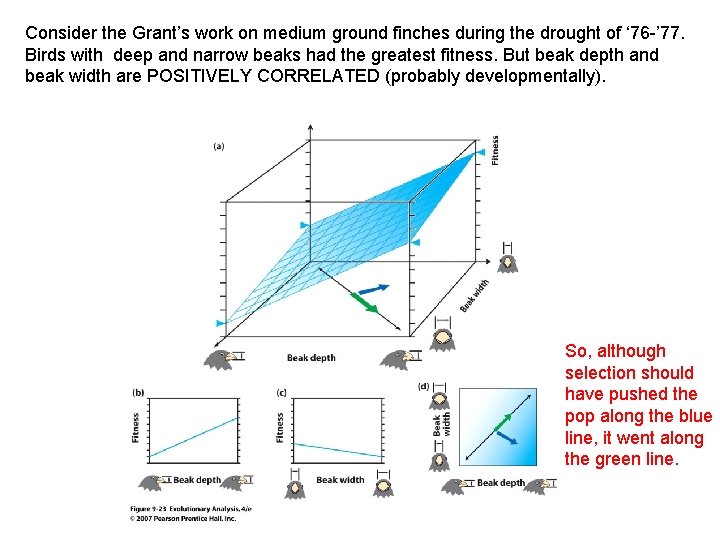
Consider the Grant’s work on medium ground finches during the drought of ‘ 76 -’ 77. Birds with deep and narrow beaks had the greatest fitness.

Consider the Grant’s work on medium ground finches during the drought of ‘ 76 -’ 77. Birds with deep and narrow beaks had the greatest fitness. But beak depth and beak width are POSITIVELY CORRELATED (probably developmentally).

Consider the Grant’s work on medium ground finches during the drought of ‘ 76 -’ 77. Birds with deep and narrow beaks had the greatest fitness. But beak depth and beak width are POSITIVELY CORRELATED (probably developmentally). So, although selection should have pushed the pop along the blue line, it went along the green line.

IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance C. Selection on Quantitative Traits D. Selection and Genetic Variation

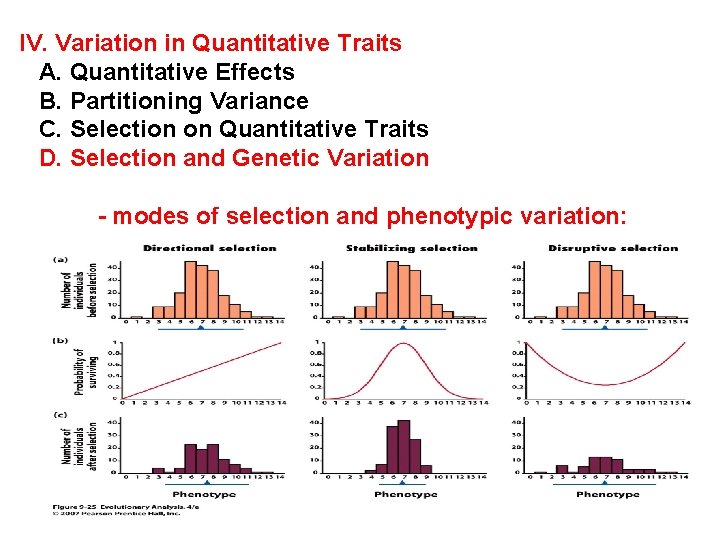
IV. Variation in Quantitative Traits A. Quantitative Effects B. Partitioning Variance C. Selection on Quantitative Traits D. Selection and Genetic Variation - modes of selection and phenotypic variation:

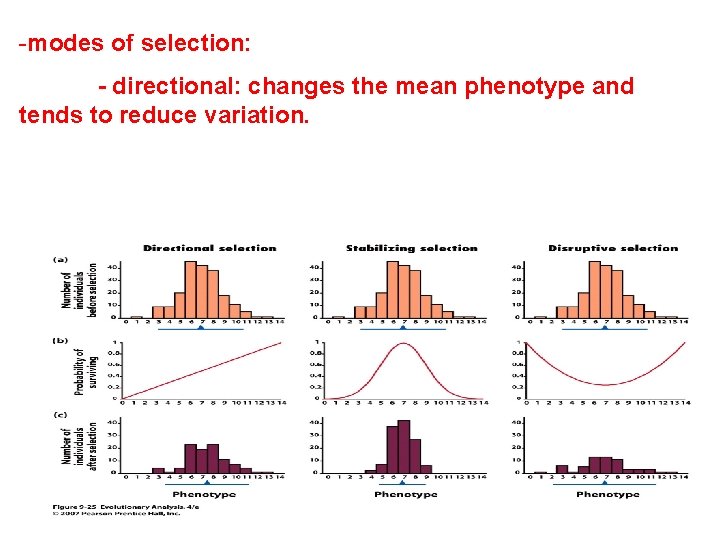
-modes of selection: - directional: changes the mean phenotype and tends to reduce variation.

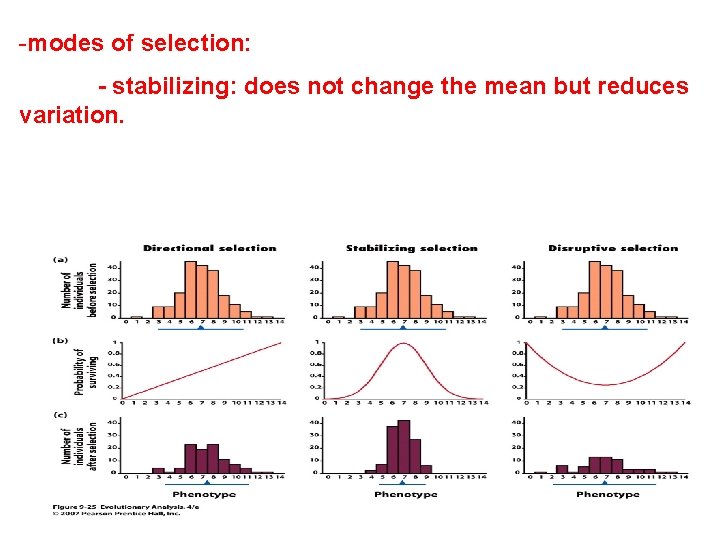
-modes of selection: - stabilizing: does not change the mean but reduces variation.

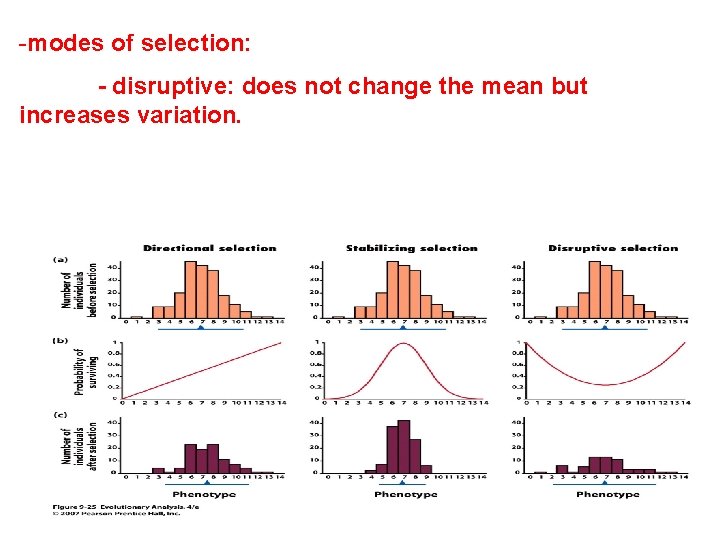
-modes of selection: - disruptive: does not change the mean but increases variation.

Directional

Stabilizing:

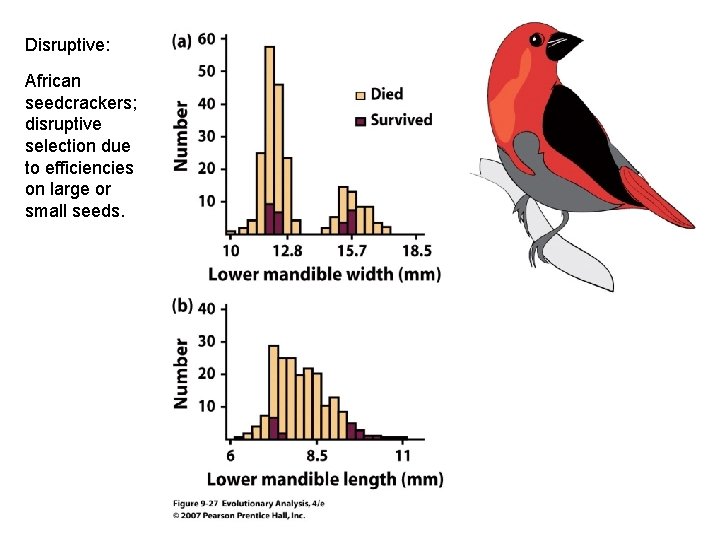
Disruptive: African seedcrackers; disruptive selection due to efficiencies on large or small seeds.

D. Selection and Genetic Variation - modes of selection and phenotypic variation: if most selection is directional and stabilizing, then variation is reduced; including genetic variation (these are quantitative traits, not single genes maintained by heterozygote advantage at one locus).

D. Selection and Genetic Variation - modes of selection and phenotypic variation: if most selection is directional and stabilizing, then variation is reduced; including genetic variation (these are quantitative traits, not single genes maintained by heterozygote advantage at one locus). But, even for very adaptive traits, there is usually still phenotypic and genetic variation. Why?

D. Selection and Genetic Variation - modes of selection and phenotypic variation: - sources of variation - new adaptive mutations are constantly produced and are increasing in frequency in the population.

D. Selection and Genetic Variation - modes of selection and phenotypic variation: - sources of variation: - new adaptive mutations are constantly produced and are increasing in frequency in the population. - deleterious mutations are maintained at low frequency; especially for genes contributing to quantitative traits where the selective pressure on any one locus may be weak.

D. Selection and Genetic Variation - modes of selection and phenotypic variation: - sources of variation: - new adaptive mutations are constantly produced and are increasing in frequency in the population. - deleterious mutations are maintained at low frequency; especially for genes contributing to quantitative traits where the selective pressure on any one locus may be weak, or recessive alleles. - disruptive, frequency dependent, multiple niche polymorphisms, etc. , in which the adaptive value of existing alleles changes through time or across space within the population.