Issues The current perception of modeling in the



- Slides: 13

Issues • The current perception of modeling in the biomedical and clinical research community, what needs to change to encourage more acceptance? • Future biomedical and clinical applications for models, based on current success stories, things that couldn't be solved w/o models, time to cure – e. g. comparative effectiveness research

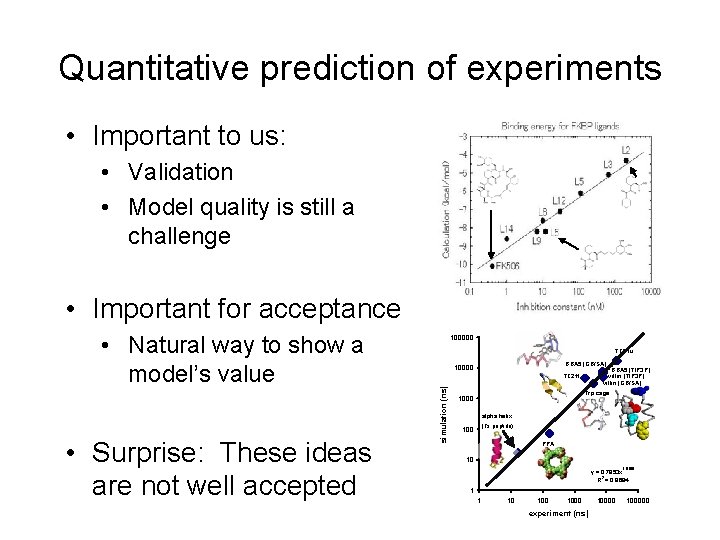
Quantitative prediction of experiments • Important to us: • Validation • Model quality is still a challenge • Important for acceptance • Surprise: These ideas are not well accepted TZ 2 tu BBA 5 (GB/SA) 10000 BBA 5 (TIP 3 P) villin (GB/SA) TZ 2 tf simulation (ns) • Natural way to show a model’s value 100000 Trp cage 1000 alpha helix (Fs peptide) 100 PPA 10 1. 0106 y = 0. 7853 x 2 R = 0. 9684 1 1 10 1000 experiment (ns) 100000

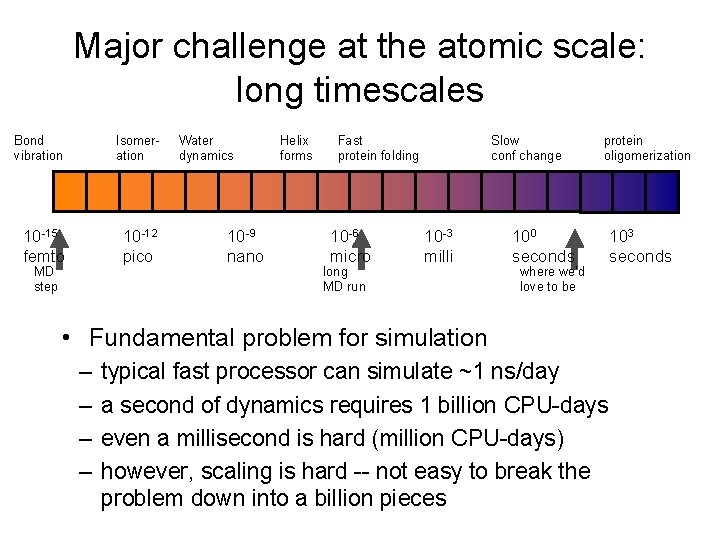
Major challenge at the atomic scale: long timescales Bond vibration Isomeration 10 -15 femto 10 -12 pico MD step Water dynamics 10 -9 nano Helix forms Fast protein folding 10 -6 micro long MD run Slow conf change 10 -3 milli protein oligomerization 100 seconds where we’d love to be • Fundamental problem for simulation – – typical fast processor can simulate ~1 ns/day a second of dynamics requires 1 billion CPU-days even a millisecond is hard (million CPU-days) however, scaling is hard -- not easy to break the problem down into a billion pieces 103 seconds

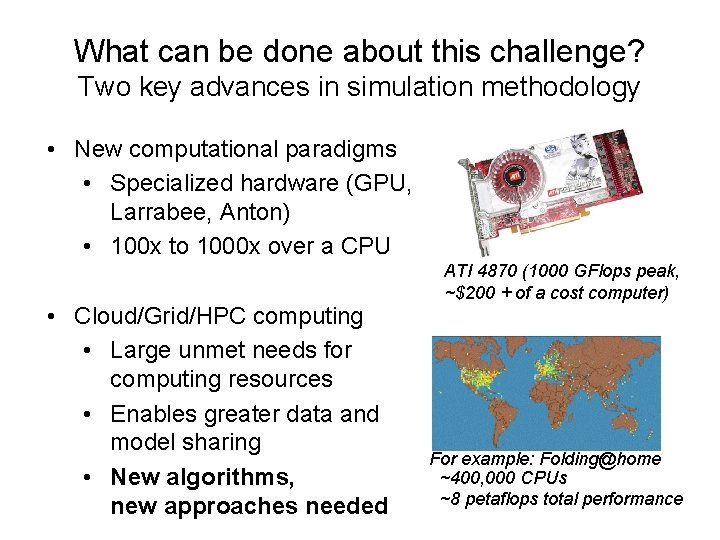
What can be done about this challenge? Two key advances in simulation methodology • New computational paradigms • Specialized hardware (GPU, Larrabee, Anton) • 100 x to 1000 x over a CPU • Cloud/Grid/HPC computing • Large unmet needs for computing resources • Enables greater data and model sharing • New algorithms, new approaches needed ATI 4870 (1000 GFlops peak, ~$200 + of a cost computer) For example: Folding@home ~400, 000 CPUs ~8 petaflops total performance

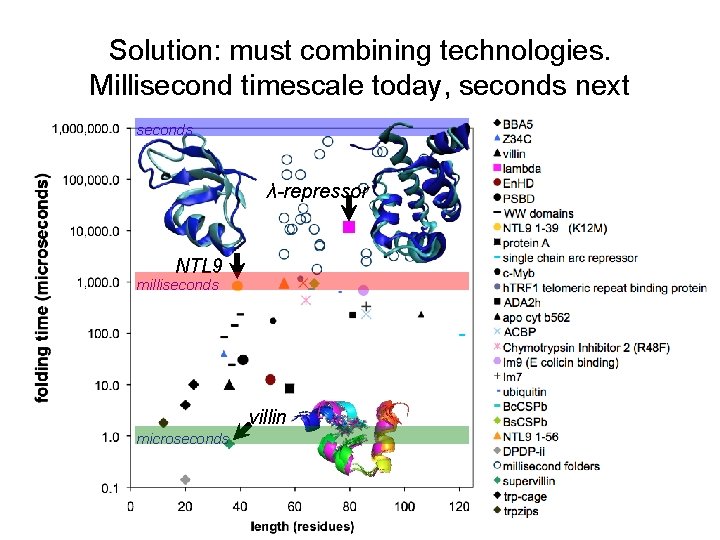
Solution: must combining technologies. Millisecond timescale today, seconds next seconds λ-repressor NTL 9 milliseconds villin microseconds

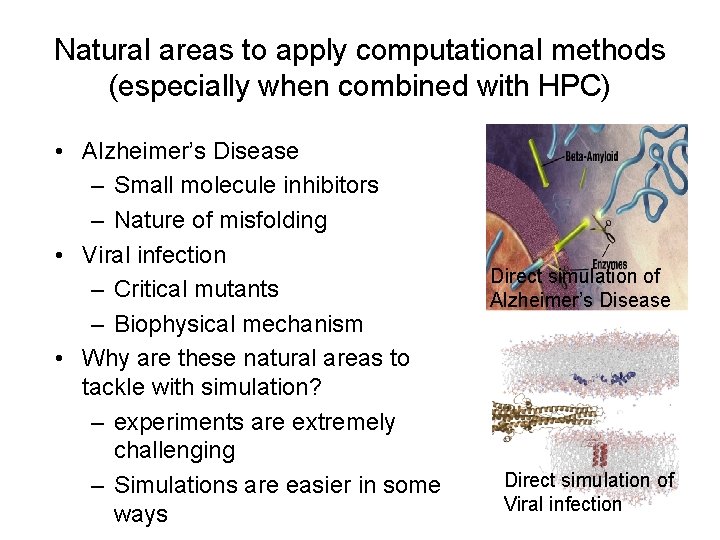
Natural areas to apply computational methods (especially when combined with HPC) • Alzheimer’s Disease – Small molecule inhibitors – Nature of misfolding • Viral infection – Critical mutants – Biophysical mechanism • Why are these natural areas to tackle with simulation? – experiments are extremely challenging – Simulations are easier in some ways Direct simulation of Alzheimer’s Disease Direct simulation of Viral infection

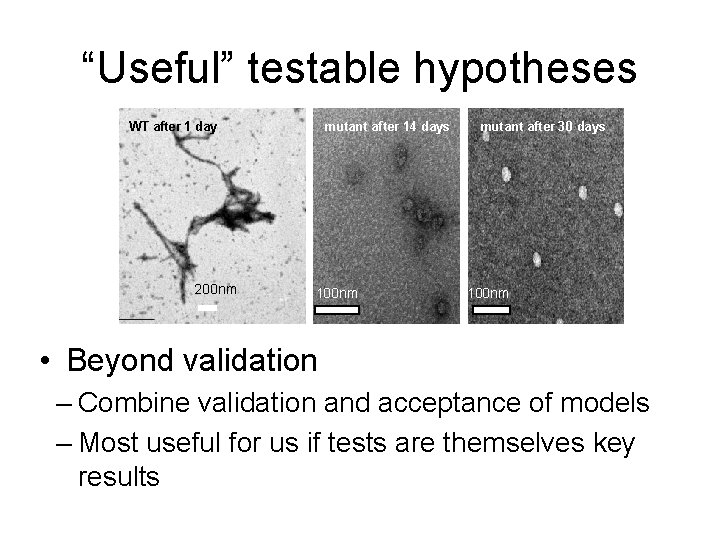
“Useful” testable hypotheses WT after 1 day 200 nm mutant after 14 days 100 nm mutant after 30 days 100 nm • Beyond validation – Combine validation and acceptance of models – Most useful for us if tests are themselves key results

For discussion • The current perception of modeling in the biomedical and clinical research community, what needs to change to encourage more acceptance? – going “over the top” in terms of connection to experiment – education on methods: many experimentalists can’t judge model quality – looking for experimentalists who need computational help

For discussion • Role of High Performance Computing (HPC) in biomedical computation – insatiable need, challenge in accessing – common approaches between scales will help in many ways (advances from one scale and transform another) – Additional traditional supercomputer resources – Ways to share distributed resources like Folding@home more broadly could transform several fields


Other success stories • computational drug design – Several labs and biotechs have examples – computation is not “all or nothing” but has become well integrated (almost silently) into many areas of drug design • protein design – inverse of protein folding: predict sequence from structure – numerous examples of successes

Broader impacts • Pharmaceutical company interest – specific collaborations (Acumen, Vertex, Numerate) • simulations driving experiments • understand limitations

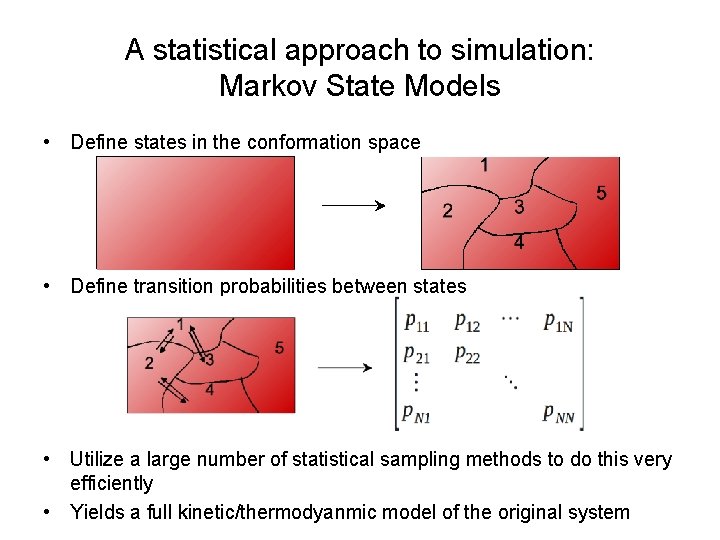
A statistical approach to simulation: Markov State Models • Define states in the conformation space • Define transition probabilities between states • Utilize a large number of statistical sampling methods to do this very efficiently • Yields a full kinetic/thermodyanmic model of the original system