Introduction to Oncomine Xiayu Stacy Huang What is




- Slides: 12

Introduction to Oncomine Xiayu Stacy Huang

What is Oncomine? • Oncomine is a cancer-specific microarray database and has a web-based data-mining platform aimed at facilitating discovery from genome-wide expression analysis • Oncomine was originally developed as an academic tool at the University of Michigan in 2003 and then became a commercial tool in 2006 (Compendia Bioscience). • Oncomine is the world’s largest existing human cancer genomic database that covers 674 independent datasets and more than 73, 000 samples.

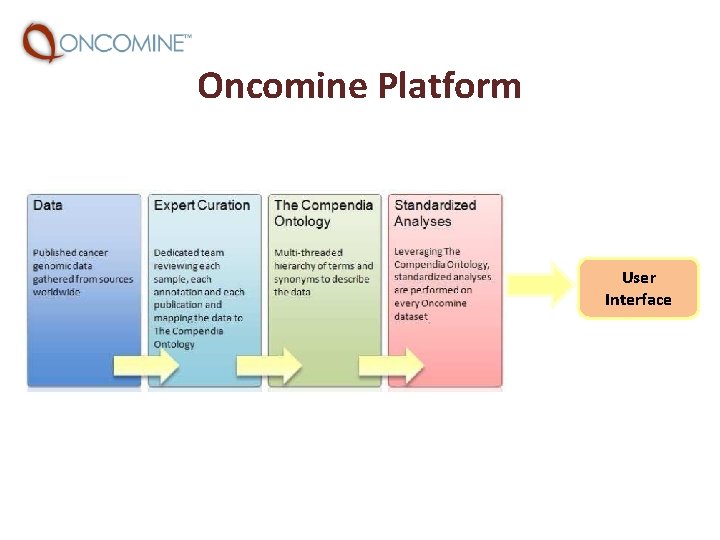
Oncomine Platform User Interface

What can Oncomine do? • Differential gene expression: What genes are over- or under - expressed in particular cancer types or subtypes? • Co-expression: Is my target gene coordinately expressed with other genes that are members of a biological pathway activated in a cancer type? • Outlier analysis: What genes might be good biomarkers for cancer subtypes? • Comparing (meta-analysis): What gene expression patterns or gene sets are validated across multiple datasets? • Concept list: Can patient subtypes be associated with this signature or gene list representing underlying biology?

Oncomine Editions • Free research edition including – Gene expression and DNA copy number data from clinical and cell-line experiments – Lists of DEGs across multiple experiments – Single gene search (differential expression, co-expression, etc) • Premium research edition including additional features – Multi-gene search – Expanded analysis types including cancer subtypes, drug sensitivity, clinical outcome analysis, etc. – Export and share results, upload custom concepts, and access commercial-level support

How to start? • Requirements – – Internet access Internet explorer 6. 0 or higher, firefox Java script must be enabled Oncomine login (https: //www. oncomine. org) • Example questions: – EGFR over-expression in brain cancer? – any genes that are coexpressed with EGFR? – EGFR expression pattern consistent across multiple studies?

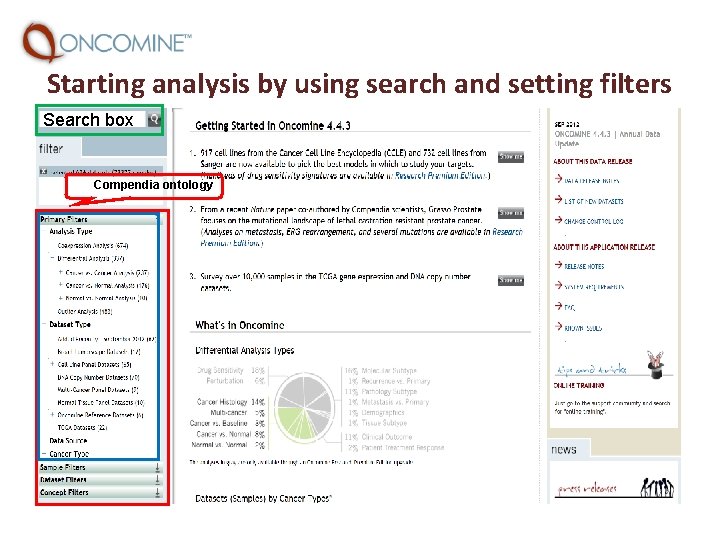
Starting analysis by using search and setting filters Search box Compendia ontology

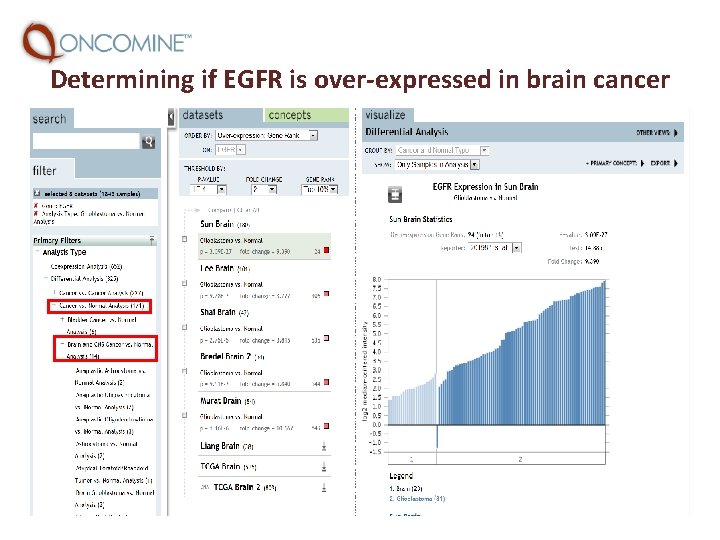
Determining if EGFR is over-expressed in brain cancer

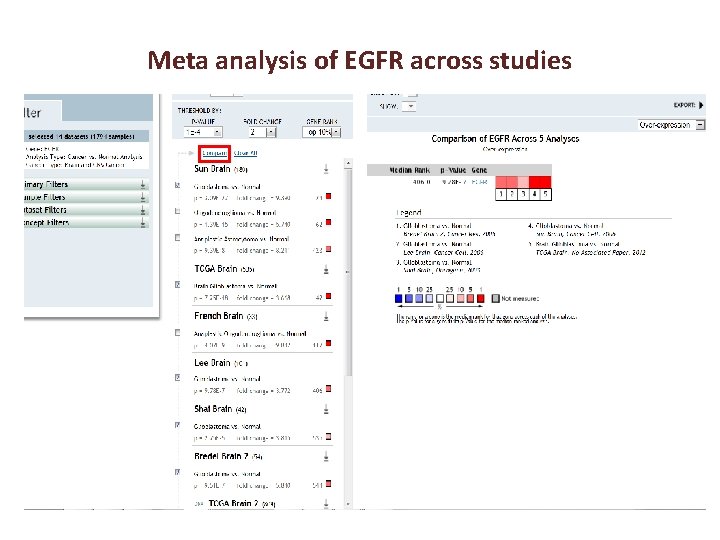
Meta analysis of EGFR across studies

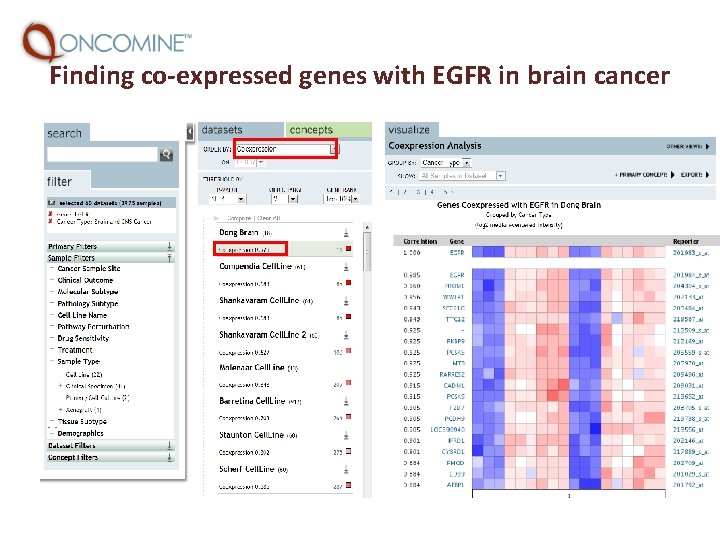
Finding co-expressed genes with EGFR in brain cancer

Conclusion • By looking at brain and CNS cancer vs. normal analyses, we can conclude EGFR is over-expressed in several independent datasets • By doing meta-analysis, we can conclude the expression pattern of EGFR is quite consistent across multiple independent studies • By doing coexpression analysis, we can conclude there are some genes that are coexpressed with EGFR and further investigation of these coexpressed genes may be needed

Thanks! Any Questions?