Introduction to Microarray Analysis and Technology Dave Lin























- Slides: 40

Introduction to Microarray Analysis and Technology Dave Lin - November 5, 2001

Overview —Why Biologists care about Genomics —Why statisticians/computer scientists —may care about genomics • Preprocessing issues • Sources of variability in constructing microarrays • Postprocessing issues • Analysis of data

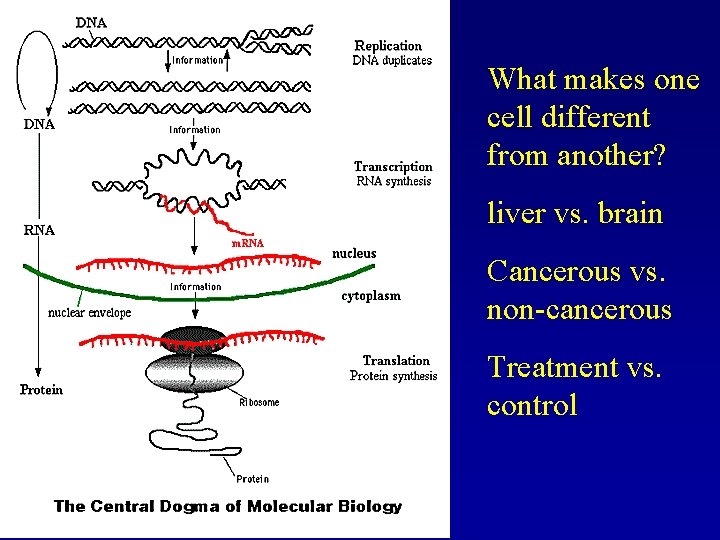
What makes one cell different from another? liver vs. brain Cancerous vs. non-cancerous Treatment vs. control

Old Days 100, 000 genes in mammalian genome each cell expresses 15, 000 of these genes each gene is expressed at a different level estimated total of 100, 000 copies of m. RNA/cell 1 -5 copies/cell - “rare” -~30% of all genes 10 -200 copies/cell - “moderate” 200 copies/cell and up - “abundant”

Cells can be defined by: Complement of Genes (which genes are expressed) How much of each gene is expressed (quantity) What makes one cell different from another? Try and find genes that are differentially expressed Study the function of these genes Find which genes interact with your favorite gene Extremely time-consuming. Huge amounts of effort expended to find individual genes that may differ between two conditions

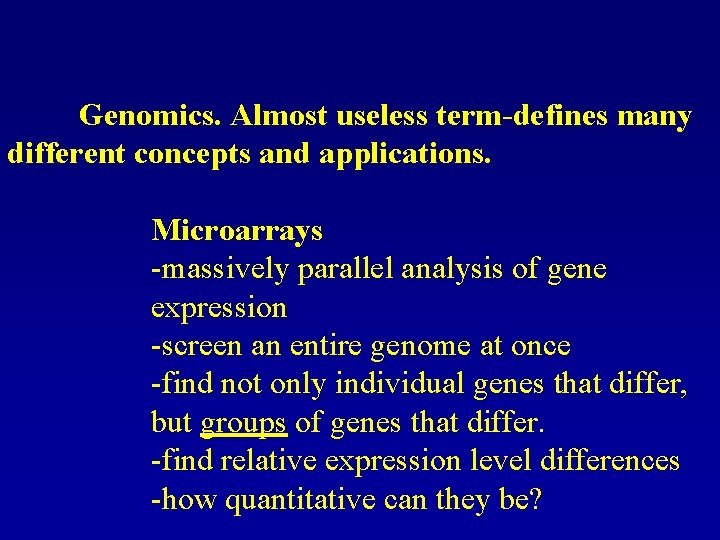
Genomics. Almost useless term-defines many different concepts and applications. Microarrays -massively parallel analysis of gene expression -screen an entire genome at once -find not only individual genes that differ, but groups of genes that differ. -find relative expression level differences -how quantitative can they be?

Microarrays. Based on old technique many flavors- majority are of two essential varieties c. DNA Arrays printing on glass slides miniaturization, throughput fluorescence based detection Affymetrix Arrays in situ synthesis of oligonucleotides will not consider Affymetrix arrays further.


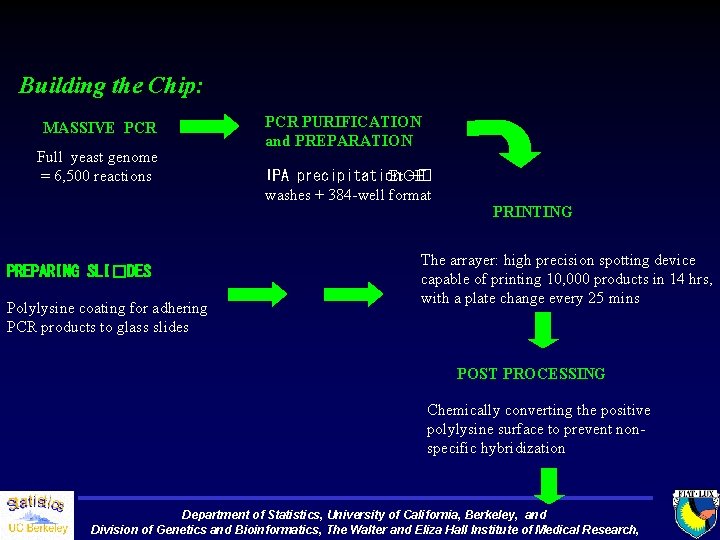
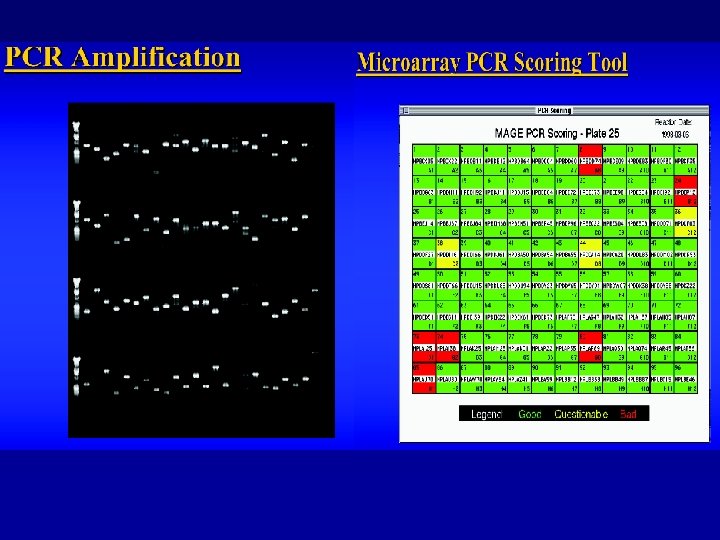
Building the Chip: MASSIVE PCR Full yeast genome = 6, 500 reactions PREPARING SLI�DES Polylysine coating for adhering PCR products to glass slides PCR PURIFICATION and PREPARATION IPA precipitation Et. OH +� washes + 384 -well format PRINTING The arrayer: high precision spotting device capable of printing 10, 000 products in 14 hrs, with a plate change every 25 mins POST PROCESSING Chemically converting the positive polylysine surface to prevent nonspecific hybridization Department of Statistics, University of California, Berkeley, and Division of Genetics and Bioinformatics, The Walter and Eliza Hall Institute of Medical Research,

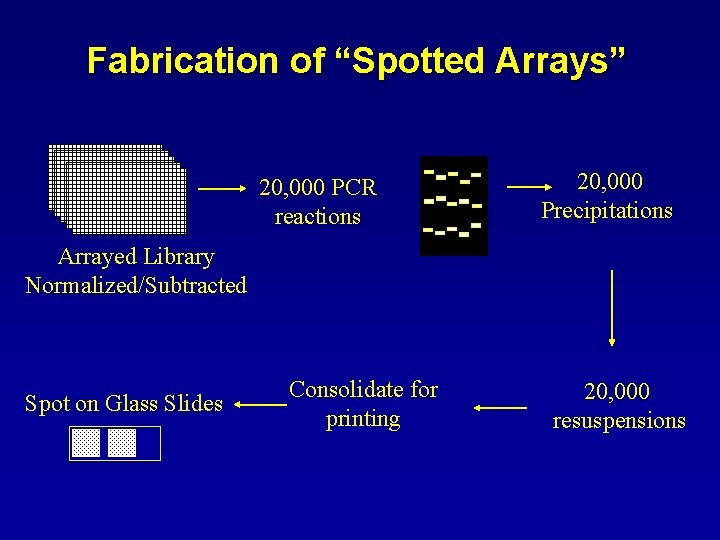
Fabrication of “Spotted Arrays” 20, 000 PCR reactions 20, 000 Precipitations Arrayed Library Normalized/Subtracted Spot on Glass Slides Consolidate for printing 20, 000 resuspensions



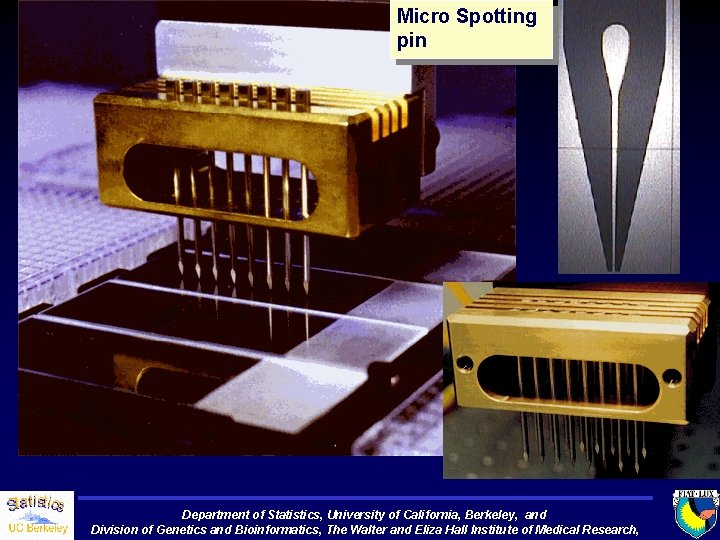
Micro Spotting pin Department of Statistics, University of California, Berkeley, and Division of Genetics and Bioinformatics, The Walter and Eliza Hall Institute of Medical Research,



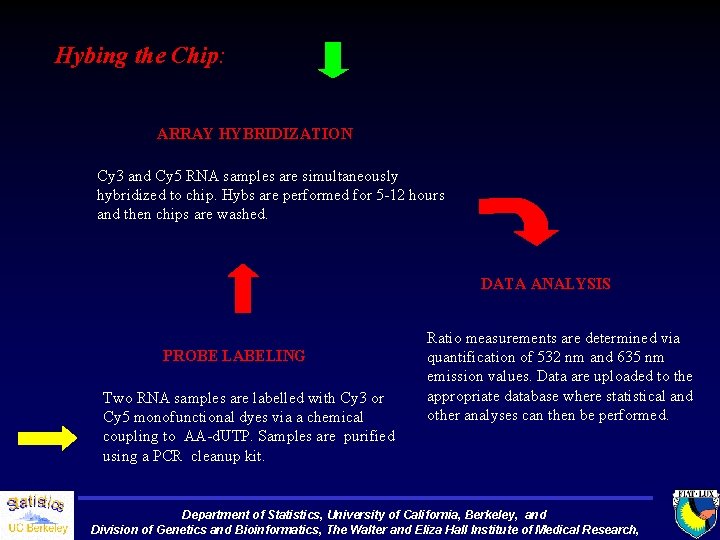
Hybing the Chip: ARRAY HYBRIDIZATION Cy 3 and Cy 5 RNA samples are simultaneously hybridized to chip. Hybs are performed for 5 -12 hours and then chips are washed. DATA ANALYSIS PROBE LABELING Two RNA samples are labelled with Cy 3 or Cy 5 monofunctional dyes via a chemical coupling to AA-d. UTP. Samples are purified using a PCR cleanup kit. Ratio measurements are determined via quantification of 532 nm and 635 nm emission values. Data are uploaded to the appropriate database where statistical and other analyses can then be performed. Department of Statistics, University of California, Berkeley, and Division of Genetics and Bioinformatics, The Walter and Eliza Hall Institute of Medical Research,

Labeling of RNAs with Cy 3 or Cy 5 Two general methods -Dye conjugated nucleotide -Amino-allyl indirect labeling

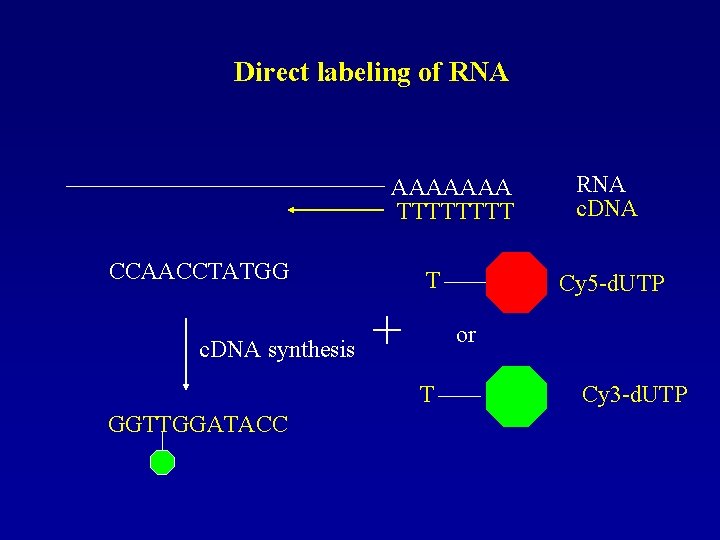
Direct labeling of RNA AAAAAAA TTTT CCAACCTATGG T c. DNA synthesis + Cy 5 -d. UTP or T GGTTGGATACC RNA c. DNA Cy 3 -d. UTP

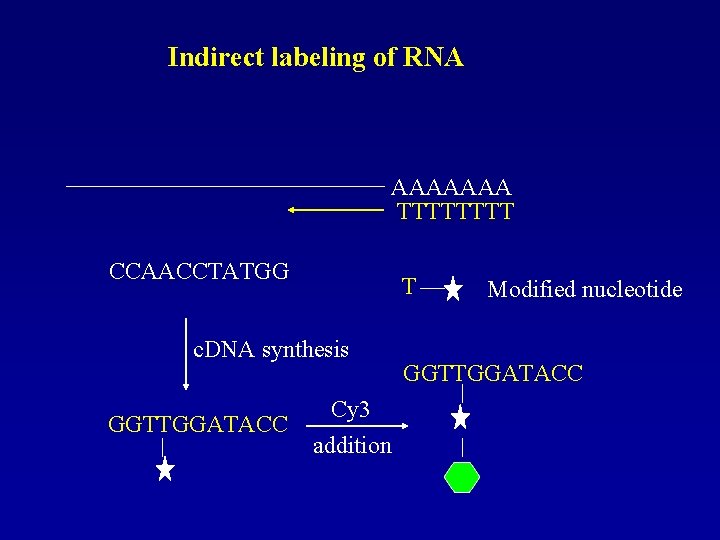
Indirect labeling of RNA AAAAAAA TTTT CCAACCTATGG T c. DNA synthesis GGTTGGATACC Cy 3 addition Modified nucleotide GGTTGGATACC

Dye effect issues Direct method Unequal incorporation of Cy 5 vs. Cy 3 Very poor overall incorporation of direct-conjugated nucleotide = more starting RNA for labeling. Indirect method Presumably less bias in initial incorporation of activated nucleotide, but not clear if more or less dye is added Both Methods Cy 3 fluoresces more brightly than Cy 5 labeling is very highly sequence dependent

Micrograph of a portion of hybridization probe from a yeast mciroarray (after hybridization). Department of Statistics, University of California, Berkeley, and Division of Genetics and Bioinformatics, The Walter and Eliza Hall Institute of Medical Research,

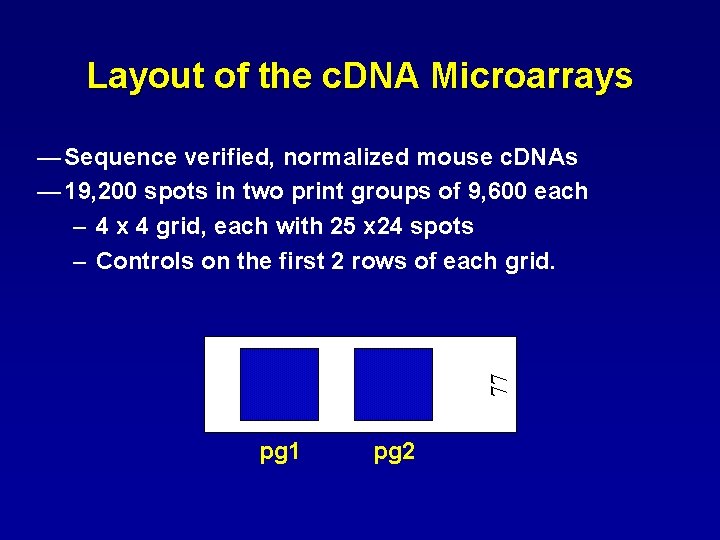
Layout of the c. DNA Microarrays 77 — Sequence verified, normalized mouse c. DNAs — 19, 200 spots in two print groups of 9, 600 each – 4 x 4 grid, each with 25 x 24 spots – Controls on the first 2 rows of each grid. pg 1 pg 2



Practical Problems 1 • Comet Tails • Likely caused by insufficiently rapid immersion of the slides in the succinic anhydride blocking solution. Department of Statistics, University of California, Berkeley , and Division of Genetics and Bioinformatics, Walter and Eliza Hall Institute of Medical Research

Practical Problems 2 Department of Statistics, University of California, Berkeley , and Division of Genetics and Bioinformatics, Walter and Eliza Hall Institute of Medical Research

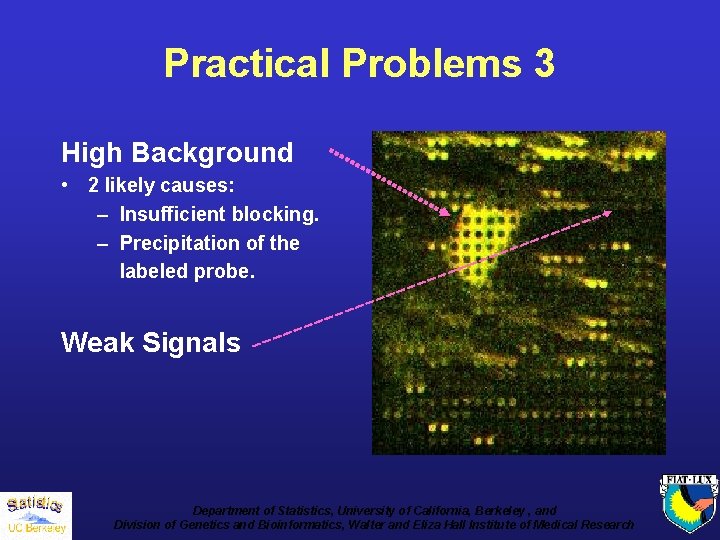
Practical Problems 3 High Background • 2 likely causes: – Insufficient blocking. – Precipitation of the labeled probe. Weak Signals Department of Statistics, University of California, Berkeley , and Division of Genetics and Bioinformatics, Walter and Eliza Hall Institute of Medical Research

Practical Problems 4 Spot overlap: Likely cause: too much rehydration during post processing. Department of Statistics, University of California, Berkeley , and Division of Genetics and Bioinformatics, Walter and Eliza Hall Institute of Medical Research

Practical Problems 5 Dust Department of Statistics, University of California, Berkeley , and Division of Genetics and Bioinformatics, Walter and Eliza Hall Institute of Medical Research


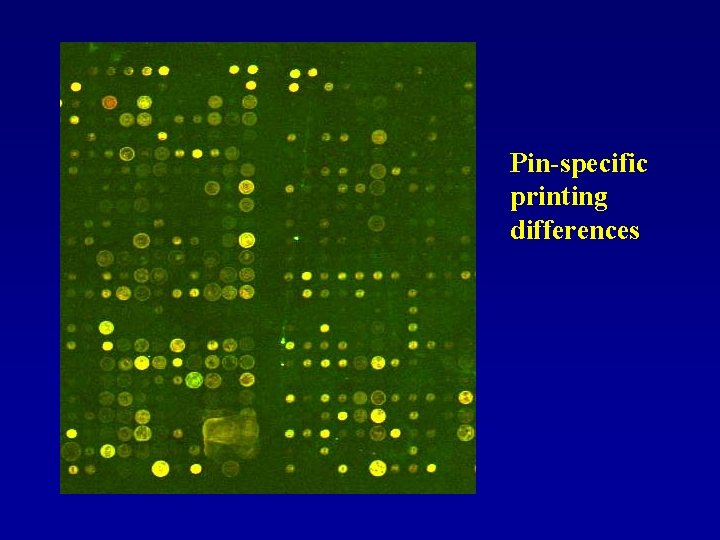
Pin-specific printing differences

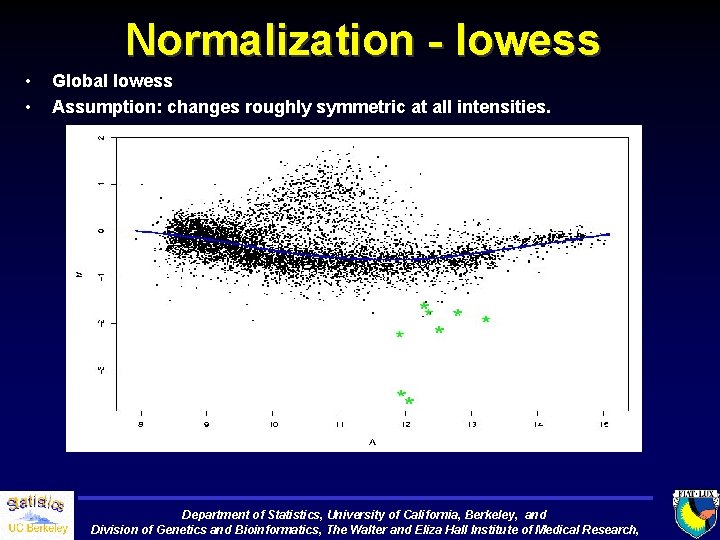
Normalization - lowess • • Global lowess Assumption: changes roughly symmetric at all intensities. Department of Statistics, University of California, Berkeley, and Division of Genetics and Bioinformatics, The Walter and Eliza Hall Institute of Medical Research,

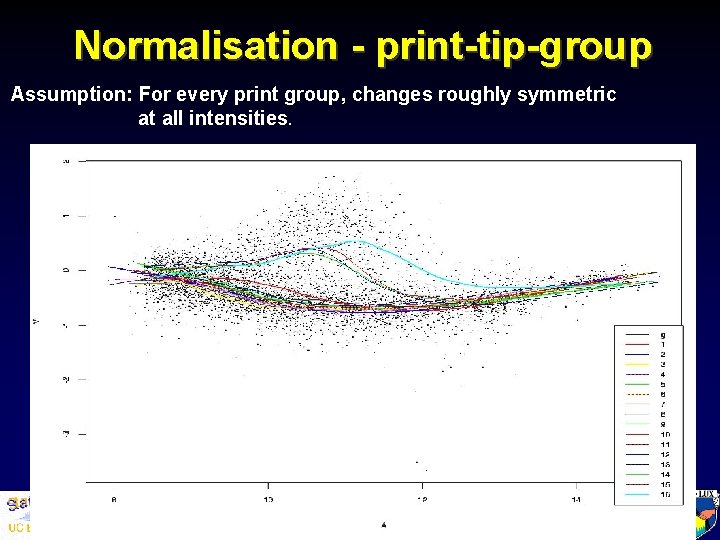
Normalisation - print-tip-group Assumption: For every print group, changes roughly symmetric at all intensities. Department of Statistics, University of California, Berkeley, and Division of Genetics and Bioinformatics, The Walter and Eliza Hall Institute of Medical Research,

Pre-processing Issues -Definition of what a real signal is what is a spot, and how to determine what should be included in the analysis? -How to determine background local (surrounding spot) vs. global (across slide) -How to correct for dye effect -How to correct for spatial effect e. g. print-tip, others -How to correct for differences between slides e. g. scale normalization

Experimental Design Issues What is the best means of performing the experiment To obtain the desired answer? Biologists’ assumptions and statisticians’ differ. Biologist viewpoint make everything exactly the same so that differences will stand out Statistician viewpoint make everything as random as possible so that real trends will stand out

Most biologists will ask- what are the differences between two samples? -implicit questions associated with microarrays. What is the best way to determine this? e. g. Design; replicates; conditions. How do I obtain the most reliable results? e. g. measurements, normalization How do I determine what a significant difference is? Do I care about “subtle” changes, or just the extremes? How is information best extracted? Is correlation useful? What type of clustering? How is information combined? How do you model the interactions of 1000 s of genes

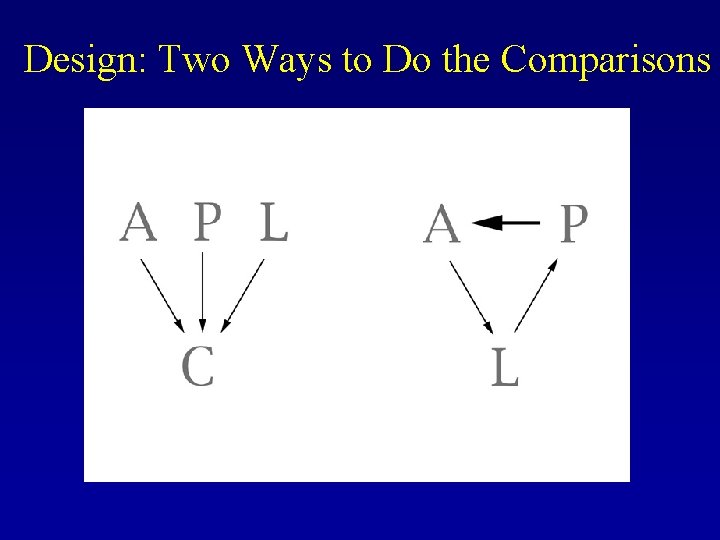
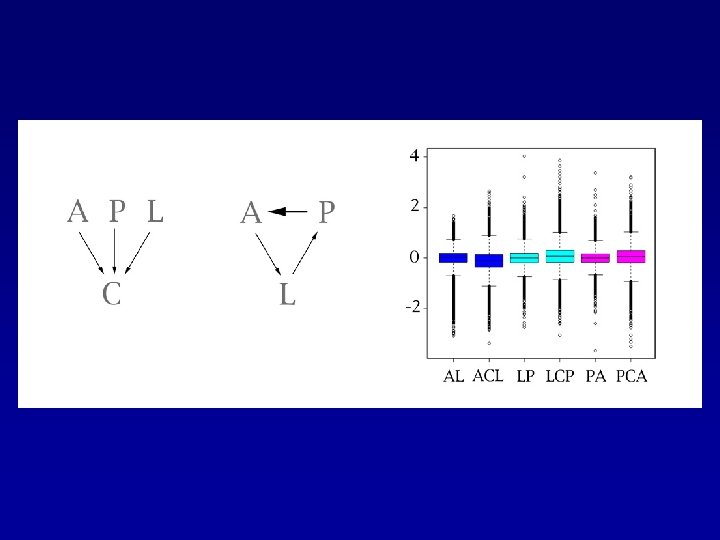
Design: Two Ways to Do the Comparisons


Advantages of Our Design —Lower variability —Increased precision —Increase in measurement of expression -> increased precision
