Introduction to Mapping of sequencing data Objectives Give

Introduction to Mapping of sequencing data
Objectives Give you an introduction to common methods used to process and analyze Next Generation Sequence data Learn methods for 1) Mapping NGS reads and 2) De novo assembly of NGS reads Give exposure to various applications for NGS experiments 2

Why? Previously we were aligning sequences in fasta files, here we align NGS sequencing data to a reference genome We may have millions of short sequences to align to a single long sequence Different approaches needed than the BLAST model of alignment 3
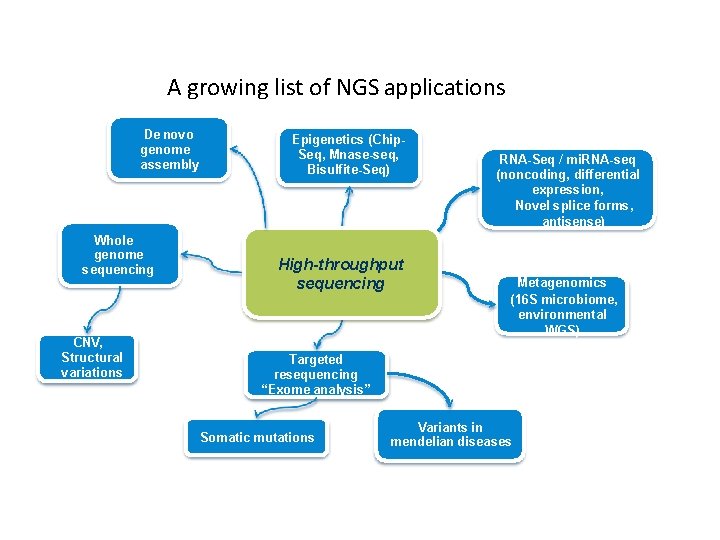
A growing list of NGS applications De novo genome assembly Whole genome sequencing CNV, Structural variations Epigenetics (Chip. Seq, Mnase-seq, Bisulfite-Seq) High-throughput sequencing RNA-Seq / mi. RNA-seq (noncoding, differential expression, Novel splice forms, antisense) Metagenomics (16 S microbiome, environmental WGS) Targeted resequencing “Exome analysis” Somatic mutations Variants in mendelian diseases
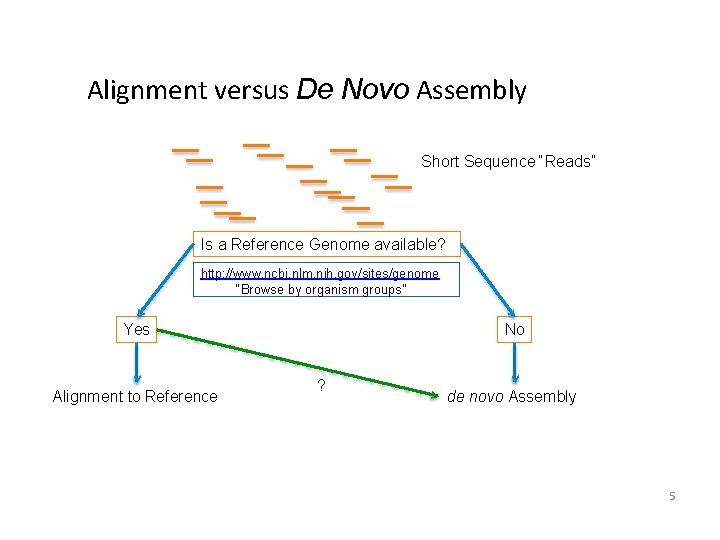
Alignment versus De Novo Assembly Short Sequence “Reads” Is a Reference Genome available? http: //www. ncbi. nlm. nih. gov/sites/genome “Browse by organism groups” Yes Alignment to Reference No ? de novo Assembly 5
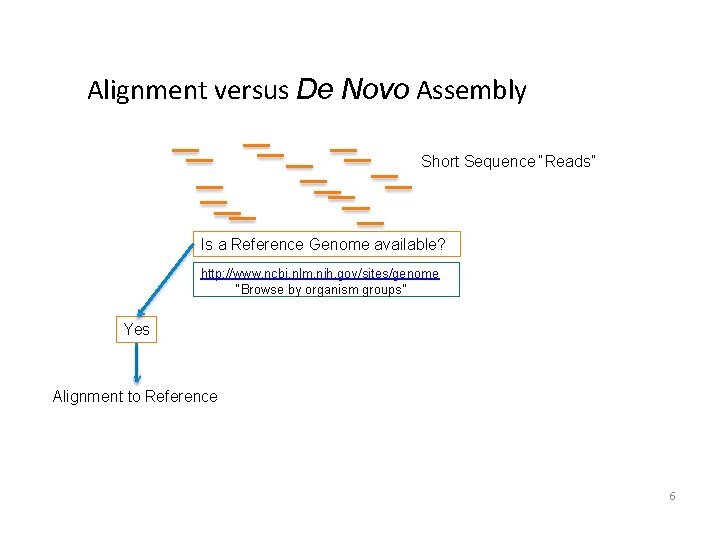
Alignment versus De Novo Assembly Short Sequence “Reads” Is a Reference Genome available? http: //www. ncbi. nlm. nih. gov/sites/genome “Browse by organism groups” Yes Alignment to Reference 6
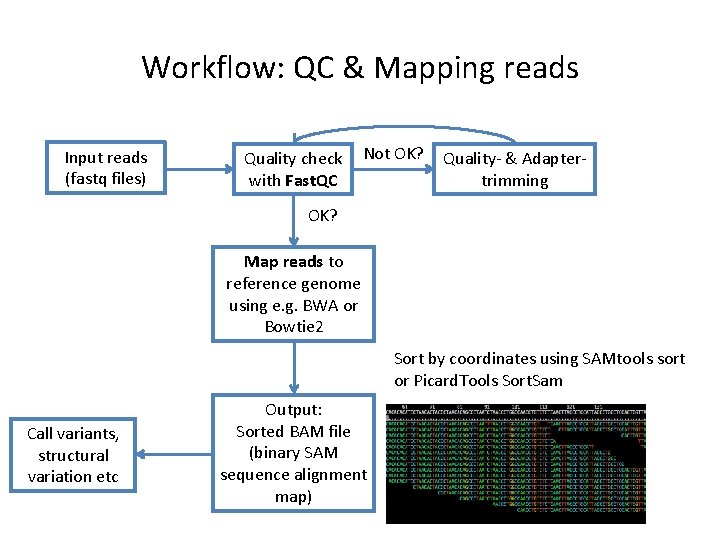
Workflow: QC & Mapping reads Input reads (fastq files) Quality check with Fast. QC Not OK? Quality- & Adaptertrimming OK? Map reads to reference genome using e. g. BWA or Bowtie 2 Sort by coordinates using SAMtools sort or Picard. Tools Sort. Sam Call variants, structural variation etc Output: Sorted BAM file (binary SAM sequence alignment map)

Short Read Alignment CTCTGCACGCGTGGGTTCGAATCCCACCTTCGTCG A chr 6 Coordinate : chr 6 27, 373, 801 8

Steps in Alignment/Mapping 1. Get your sequence data 2. Check quality of sequence data 3. Choose an alignment/mapping program 4. Run the alignment 5. View the alignments 6. Downstream Processing 9

Steps in Alignment/Mapping 1. Get your sequence data 2. Check quality of sequence data 3. Choose an alignment/mapping program 4. Run the alignment 5. View the alignments 6. Downstream Processing 10

Public Short Read Repositories NIH/NCBI • Short Read Archive (http: //www. ncbi. nlm. nih. gov/sra) • Gene Expression Omnibus (http: //www. ncbi. nlm. nih. gov/geo/) • 1000 Genomes (ftp: //ftp-trace. ncbi. nih. gov/1000 genomes/ftp/) • European Nucleotide Archive (http: //www. ebi. ac. uk/ena/) fastq-dump SRR 036642 11

Sequence file formats Next gen sequence file formats are based on the commonly used FASTA format >sequence_ID and optional comments ATTCCGGTGCGGTGCTGCCGGTG C TTCGAAATTGGCGTCAGT The Phred quality scores per sequenced base were added to form the FASTQ format 12
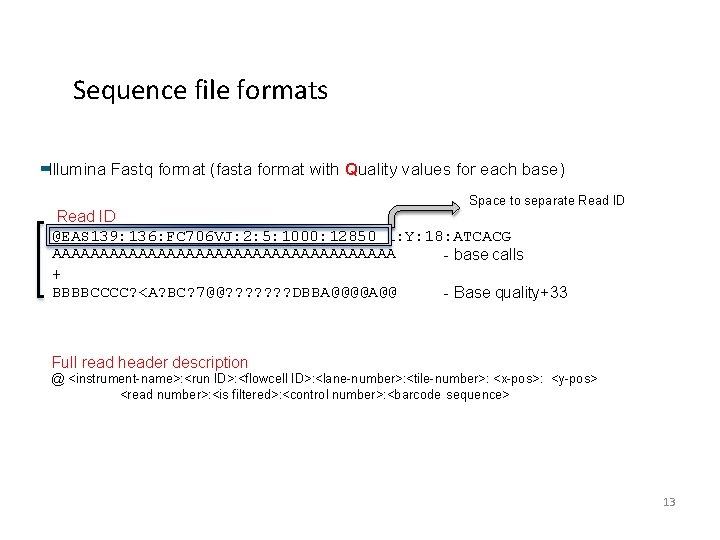
Sequence file formats Illumina Fastq format (fasta format with Quality values for each base) Space to separate Read ID @EAS 139: 136: FC 706 VJ: 2: 5: 1000: 12850 1: Y: 18: ATCACG AAAAAAAAAAAAAAAAAA - base calls + BBBBCCCC? <A? BC? 7@@? ? ? ? DBBA@@@@A@@ - Base quality+33 Full read header description @ <instrument-name>: <run ID>: <flowcell ID>: <lane-number>: <tile-number>: <x-pos>: <y-pos> <read number>: <is filtered>: <control number>: <barcode sequence> 13

Steps in Alignment/Mapping 1. Get your sequence data 2. Check quality of sequence data 3. Choose an alignment/mapping program 4. Run the alignment 5. View the alignments 6. Downstream Processing 14
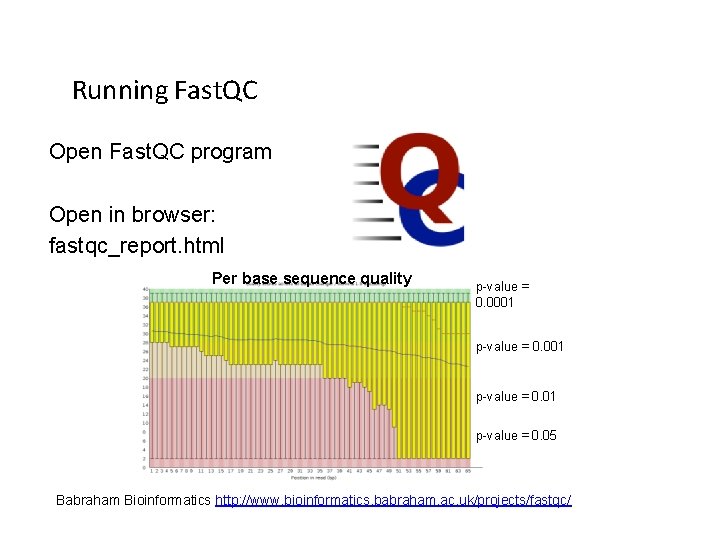
Running Fast. QC Open Fast. QC program Open in browser: fastqc_report. html Per base sequence quality p-value = 0. 0001 p-value = 0. 05 18 Babraham Bioinformatics http: //www. bioinformatics. babraham. ac. uk/projects/fastqc/

Steps in Alignment/Mapping 1. Get your sequence data 2. Check quality of sequence data 3. Choose an alignment/mapping program 4. Run the alignment 5. View the alignments 6. Downstream Processing 16
Short Read Alignment Software BFAST BLASTN BLAT Bowtie BWA ELAND GNUMAP GMAP and GSNAP MAQ mr. FAST and mrs. FAST MOSAIK Novoalign RUM SHRi. MP SOAP Splice. Map SSAHA and SSAHA 2 STAR Top. Hat ~20 more… http: //en. wikipedia. org/wiki/List_of_sequence_alignment_software http: //tinyurl. com/seqanswers-mapping 17
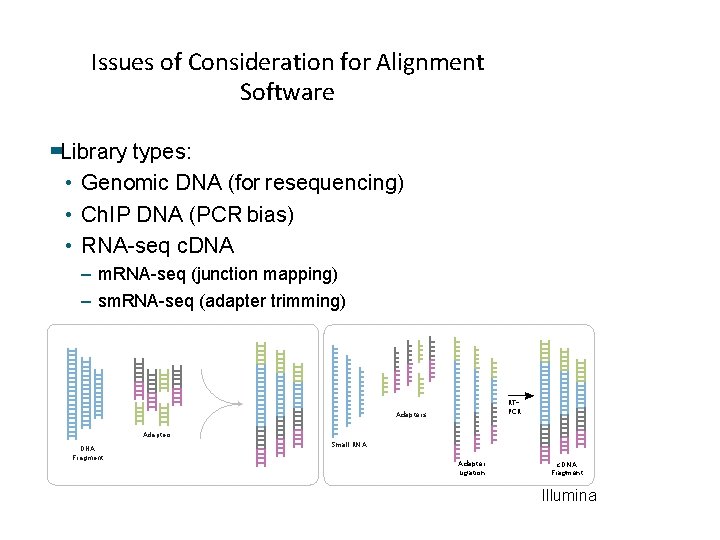
Issues of Consideration for Alignment Software Library types: • Genomic DNA (for resequencing) • Ch. IP DNA (PCR bias) • RNA-seq c. DNA – m. RNA-seq (junction mapping) – sm. RNA-seq (adapter trimming) RTPCR Adapters DNA Fragment Small RNA Adapter Ligation c. DNA Fragment Illumina 21
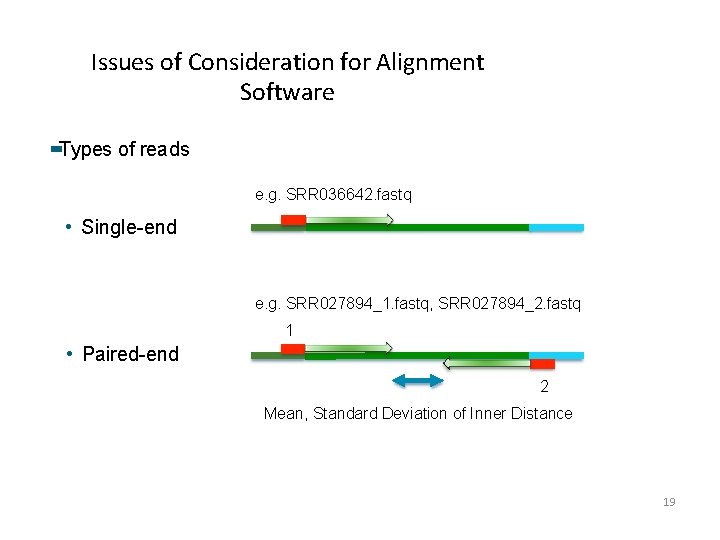
Issues of Consideration for Alignment Software Types of reads e. g. SRR 036642. fastq • Single-end e. g. SRR 027894_1. fastq, SRR 027894_2. fastq 1 • Paired-end 2 Mean, Standard Deviation of Inner Distance 19
Issues of Consideration for Alignment Software Platform differences • Bases (ACTG) • Colorspace (2 -base encoding, SOLi. D) • Read Length • 454 (homopolymers) 20
Issues of Consideration for Alignment Software Properties • Open-source or proprietary ($) • Accuracy • Speed of algorithm • Multi-threaded or single processor • RAM requirements (2 GB vs 50 GB for loading index) • Use of base quality score • Gapped alignment (indels) 21

Steps in Alignment/Mapping 1. Get your sequence data 2. Check quality of sequence data 3. Choose an alignment/mapping program 4. Run the alignment 5. View the alignments 6. Downstream Processing 22
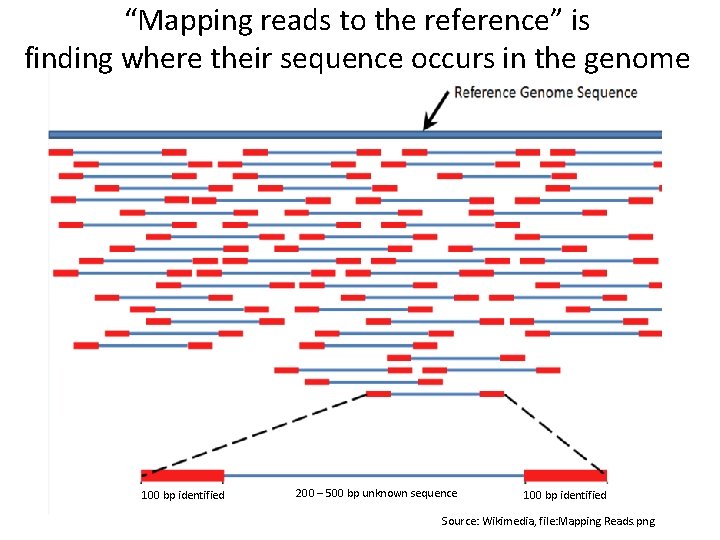
“Mapping reads to the reference” is finding where their sequence occurs in the genome 100 bp identified 200 – 500 bp unknown sequence 100 bp identified Source: Wikimedia, file: Mapping Reads. png
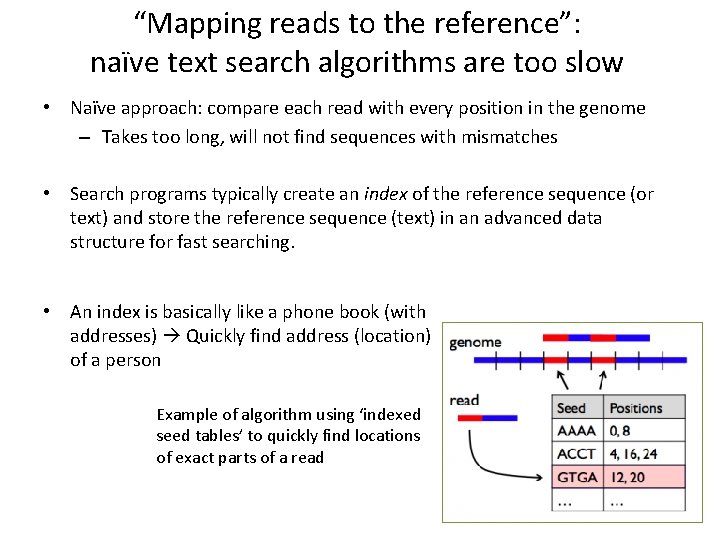
“Mapping reads to the reference”: naïve text search algorithms are too slow • Naïve approach: compare each read with every position in the genome – Takes too long, will not find sequences with mismatches • Search programs typically create an index of the reference sequence (or text) and store the reference sequence (text) in an advanced data structure for fast searching. • An index is basically like a phone book (with addresses) Quickly find address (location) of a person Example of algorithm using ‘indexed seed tables’ to quickly find locations of exact parts of a read
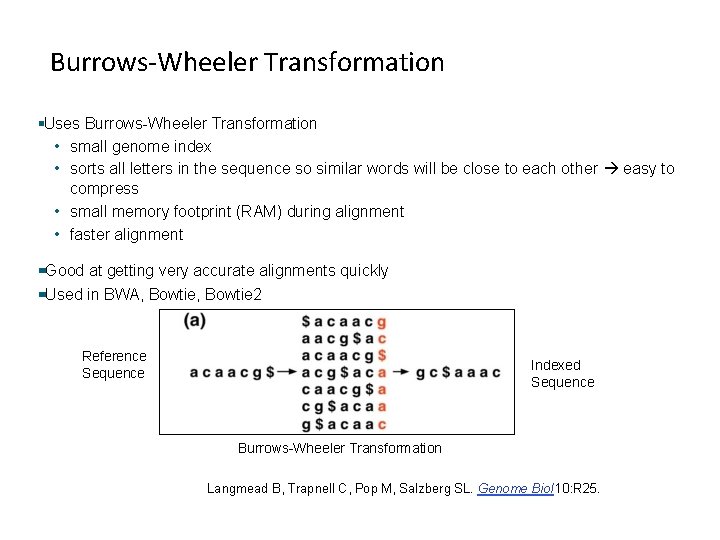
Burrows-Wheeler Transformation Uses Burrows-Wheeler Transformation • small genome index • sorts all letters in the sequence so similar words will be close to each other easy to compress • small memory footprint (RAM) during alignment • faster alignment Good at getting very accurate alignments quickly Used in BWA, Bowtie 2 Reference Sequence Indexed Sequence Burrows-Wheeler Transformation Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol 10: R 25. 36

Read Mapping: General problems • Read can match equally well at more than one location (e. g. repeats, pseudo-genes) • Reads can have imperfect hit to it’s actual position, e. g. if it carries a break point, SNP, insertion and/or deletion compared to the reference sequence

Output of read mapping: SAM and BAM files SAM = Sequence Alignment Map BAM = Binary SAM = compressed SAM Sequence Alignment/Map format contains information about how sequence reads map to a reference genome • Supports paired-end reads and color space from SOLi. D. • Is produced by bowtie, BWA and other mapping tools • •
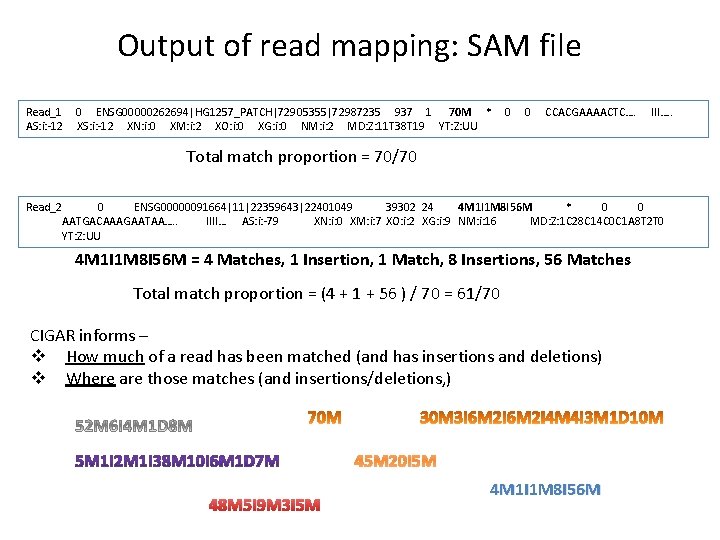
Output of read mapping: SAM file Read_1 AS: i: -12 0 ENSG 00000262694|HG 1257_PATCH|72905355|72987235 937 1 70 M * XS: i: -12 XN: i: 0 XM: i: 2 XO: i: 0 XG: i: 0 NM: i: 2 MD: Z: 11 T 38 T 19 YT: Z: UU 0 0 CCACGAAAACTC…. III…. . Total match proportion = 70/70 Read_2 0 ENSG 00000091664|11|22359643|22401049 39302 24 4 M 1 I 1 M 8 I 56 M * 0 0 AATGACAAAGAATAA…. . IIII… AS: i: -79 XN: i: 0 XM: i: 7 XO: i: 2 XG: i: 9 NM: i: 16 MD: Z: 1 C 28 C 14 C 0 C 1 A 8 T 2 T 0 YT: Z: UU 4 M 1 I 1 M 8 I 56 M = 4 Matches, 1 Insertion, 1 Match, 8 Insertions, 56 Matches Total match proportion = (4 + 1 + 56 ) / 70 = 61/70 CIGAR informs – v How much of a read has been matched (and has insertions and deletions) v Where are those matches (and insertions/deletions, ) 45 M 20 I 5 M 48 M 5 I 9 M 3 I 5 M 4 M 1 I 1 M 8 I 56 M

Harvesting Information from SAM • Query name, QNAME (SAM) / read_name (BAM). • FLAG provides the following information: – – – – – are there multiple fragments? are all fragments properly aligned? is this fragment unmapped? is the next fragment unmapped? is this query the reverse strand? is the next fragment the reverse strand? is the last fragment? is this a secondary alignment? did this read fail quality controls? is this read a PCR or optical duplicate Source: www. cs. colostate. edu/~cs 680/Slides/lecture 3. pdf

Steps in Alignment/Mapping 1. Get your sequence data 2. Check quality of sequence data 3. Choose an alignment/mapping program 4. Run the alignment 5. View the alignments 6. Downstream Processing
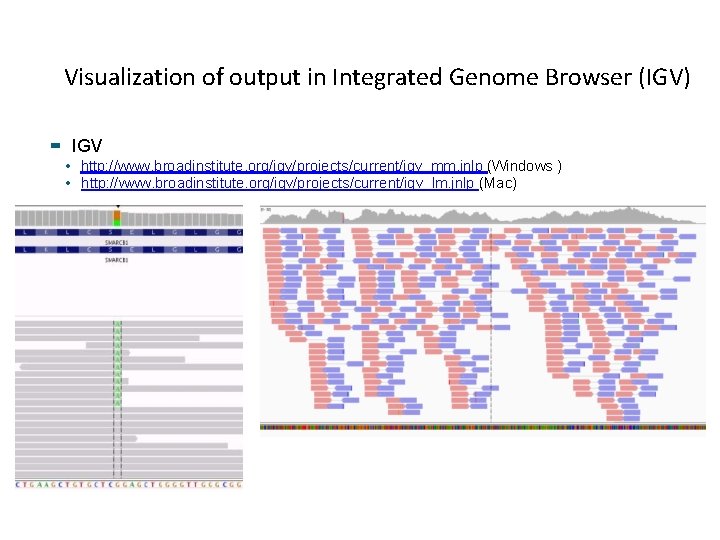
Visualization of output in Integrated Genome Browser (IGV) IGV • http: //www. broadinstitute. org/igv/projects/current/igv_mm. jnlp (Windows ) • http: //www. broadinstitute. org/igv/projects/current/igv_lm. jnlp (Mac)

Steps in Alignment/Mapping 1. Get your sequence data 2. Check quality of sequence data 3. Choose an alignment/mapping program 4. Run the alignment 5. View the alignments 6. Downstream Processing
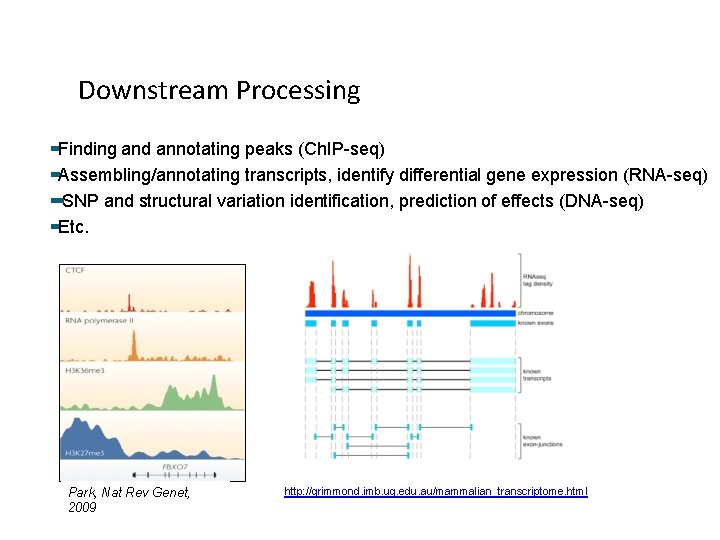
Downstream Processing Finding and annotating peaks (Ch. IP-seq) Assembling/annotating transcripts, identify differential gene expression (RNA-seq) SNP and structural variation identification, prediction of effects (DNA-seq) Etc. Park, Nat Rev Genet, 2009 http: //grimmond. imb. uq. edu. au/mammalian_transcriptome. html
Short Read Alignment: Focus on BWA 3 4
SE. Seeding and re-seeding • BWA-MEM follows the canonical seed-and-extend paradigm. • Seed an alignment with MEMs (Maximal Exact Matches), which essentially finds at each query position the longest exact match covering the position. A MEM is a match that cannot be extended in either direction of the match. • If a read cannot be aligned by extension of the MEM, reseeding is used to generate new seeds for mapping and extension.

SE. Chaining and chain filtering • We call a group of seeds that are colinear and close to each other as a chain. • We greedily chain the seeds while seeding and then filter out short chains that are largely contained in a long chain and are much worse than the long chain (by default, both 50% and 38 bp shorter than the long chain). • Chain filtering aims to reduce unsuccessful seed extension at a later step. • Chains detected here do not need to be accurate.

SE. Seed extension • rank a seed by length of the chain it belongs to and then by the seed length. • drop the seed if it is already contained in an alignment found before, or extend the seed with a banded affine-gap-penalty dynamic programming (DP) if it potentially leads to a new alignment.

Running Operation • MEM mode
Now we try using BWA-MEM to perform a mapping alignment • Working in the model plant Arabidopsis – Diploid – Good genome resources – – 120 Mbp genome 5 Chromosomes We will look at chromosome 1 only (30 Mbp) We will compare alignments from the reference variety of Arabidopsis Columbia (Col-0) to a wild variety Seattle-0
- Slides: 39