Introduction to genome annotation practical information Some possibilities
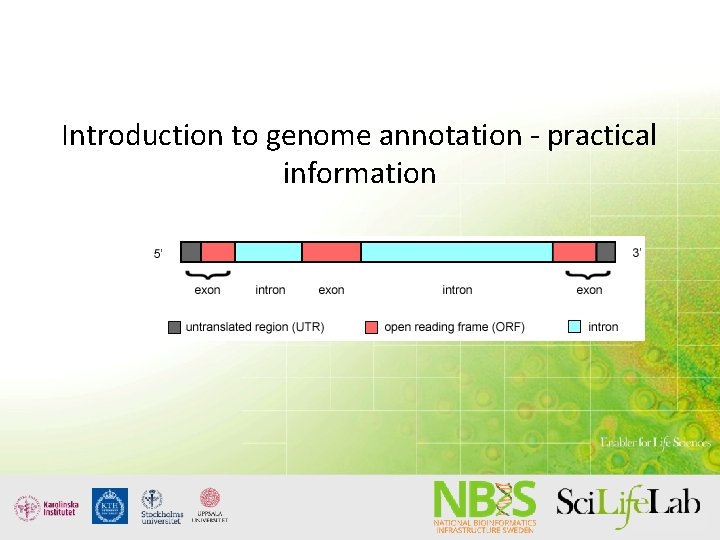
Introduction to genome annotation - practical information Some possibilities and some pitfalls

Practical info • Coffee breaks • Lunch • Dinner at Koh Phangan 18. 00
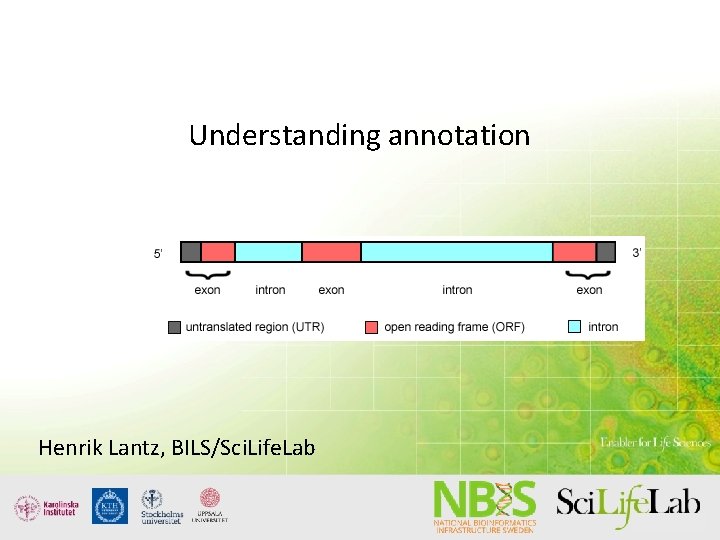
Understanding annotation Some possibilities and some pitfalls Henrik Lantz, BILS/Sci. Life. Lab
Lecture synopsis • • • What is annotation? Structural genome annotation Types of data used Transcriptome annotation Functional annotation
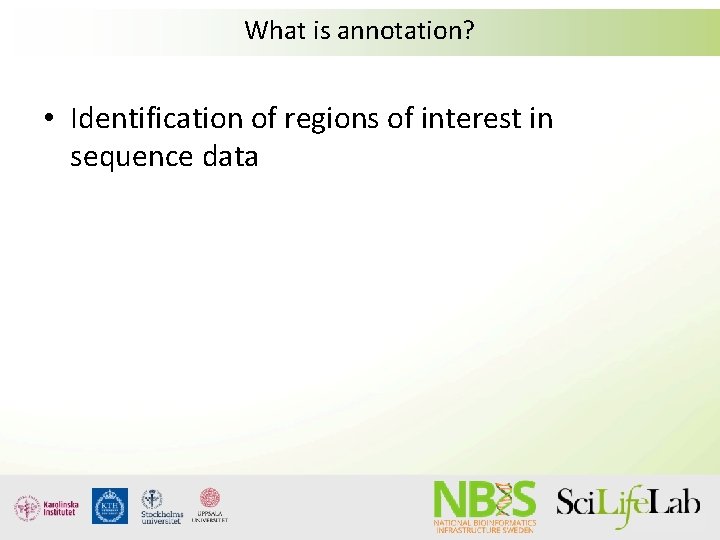
What is annotation? • Identification of regions of interest in sequence data

From a genome…
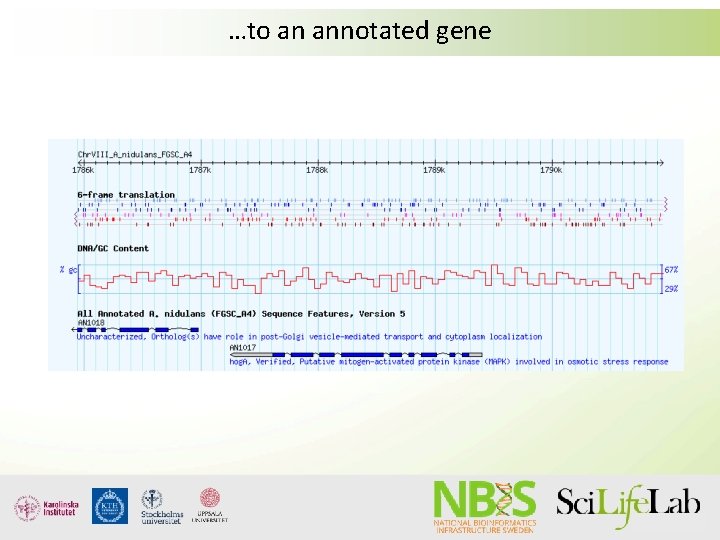
…to an annotated gene
GFF file format
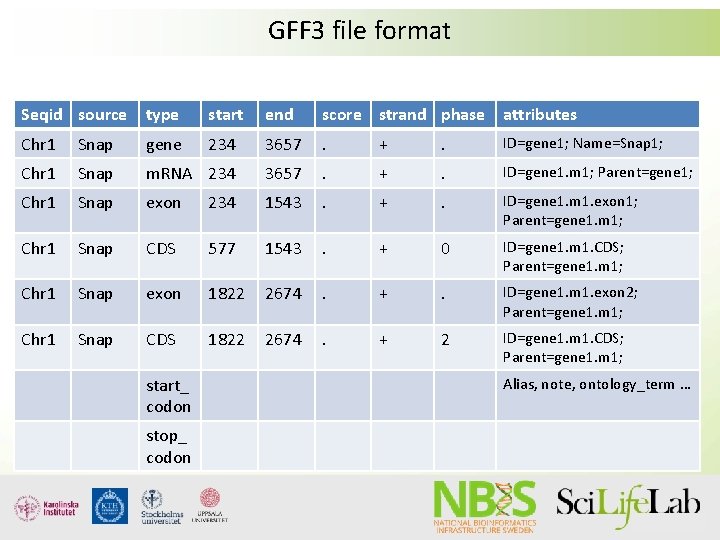
GFF 3 file format Seqid source type start end score strand phase attributes Chr 1 Snap gene 234 3657 . + . ID=gene 1; Name=Snap 1; Chr 1 Snap m. RNA 234 3657 . + . ID=gene 1. m 1; Parent=gene 1; Chr 1 Snap exon 234 1543 . + . ID=gene 1. m 1. exon 1; Parent=gene 1. m 1; Chr 1 Snap CDS 577 1543 . + 0 ID=gene 1. m 1. CDS; Parent=gene 1. m 1; Chr 1 Snap exon 1822 2674 . + . ID=gene 1. m 1. exon 2; Parent=gene 1. m 1; Chr 1 Snap CDS 1822 2674 . + 2 ID=gene 1. m 1. CDS; Parent=gene 1. m 1; start_ codon stop_ codon Alias, note, ontology_term …
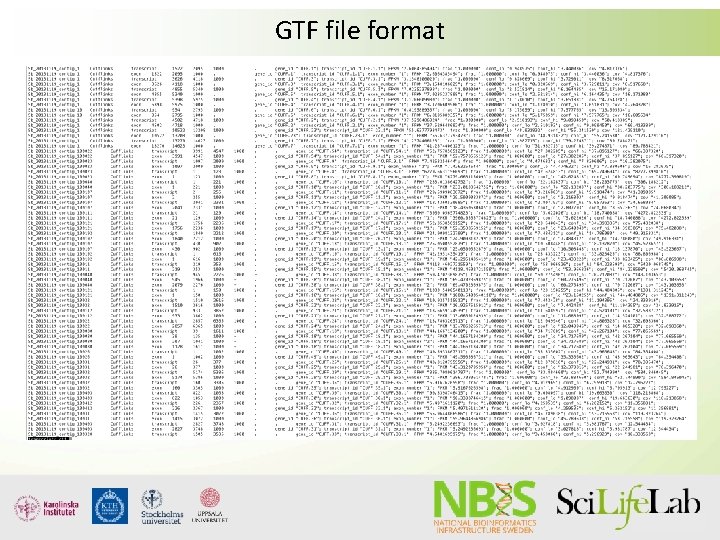
GTF file format
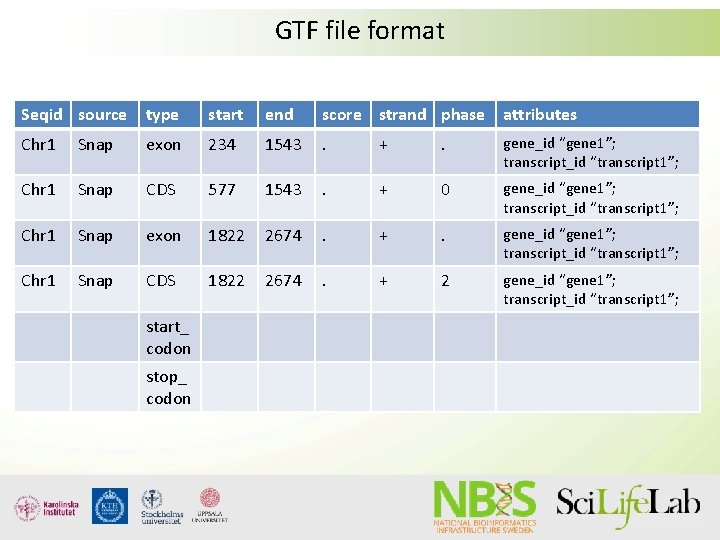
GTF file format Seqid source type start end score strand phase attributes Chr 1 Snap exon 234 1543 . + . gene_id “gene 1”; transcript_id “transcript 1”; Chr 1 Snap CDS 577 1543 . + 0 gene_id “gene 1”; transcript_id “transcript 1”; Chr 1 Snap exon 1822 2674 . + . gene_id “gene 1”; transcript_id “transcript 1”; Chr 1 Snap CDS 1822 2674 . + 2 gene_id “gene 1”; transcript_id “transcript 1”; start_ codon stop_ codon
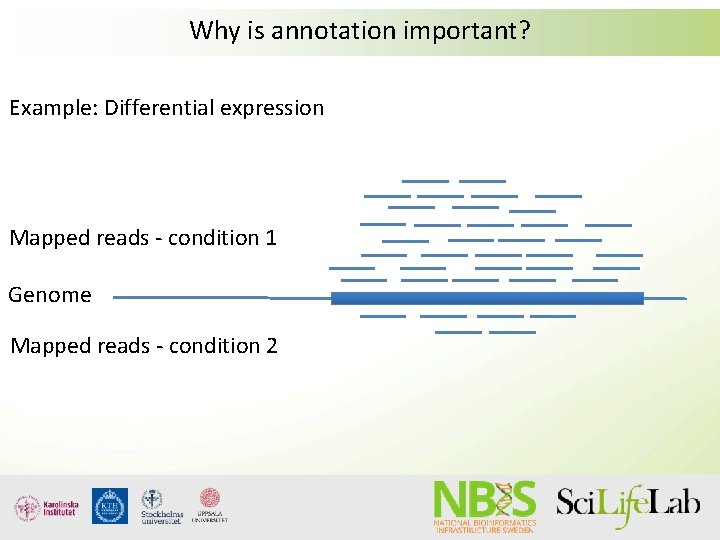
Why is annotation important? Example: Differential expression Mapped reads - condition 1 Genome Mapped reads - condition 2
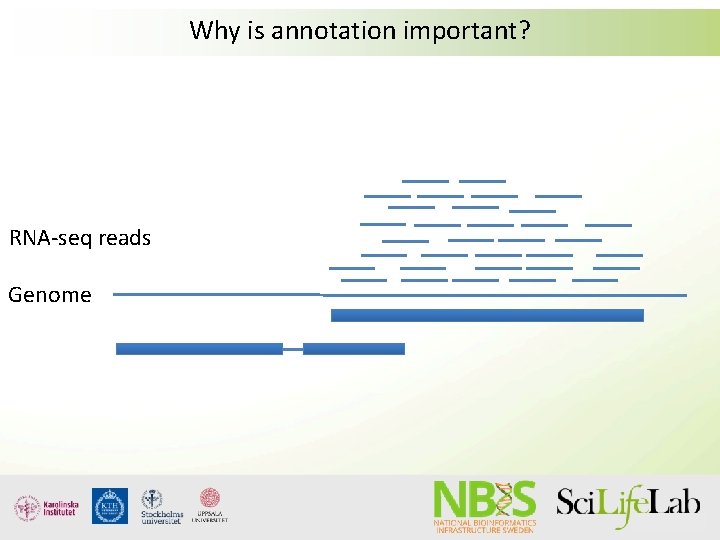
Why is annotation important? RNA-seq reads Genome
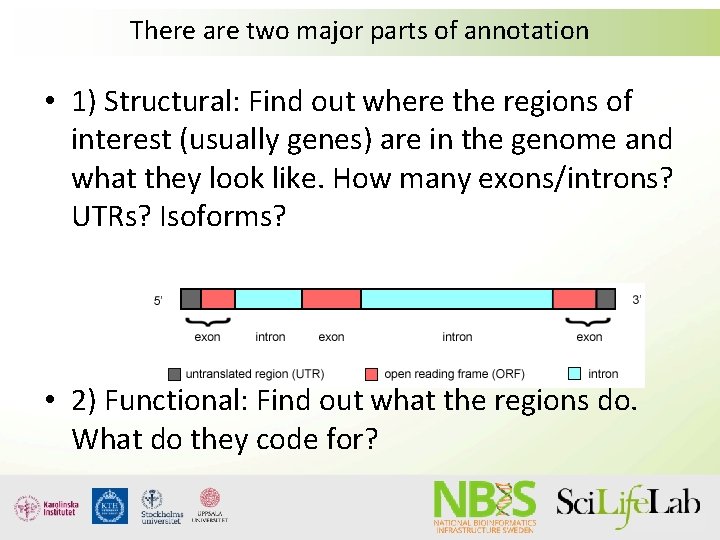
There are two major parts of annotation • 1) Structural: Find out where the regions of interest (usually genes) are in the genome and what they look like. How many exons/introns? UTRs? Isoforms? • 2) Functional: Find out what the regions do. What do they code for?
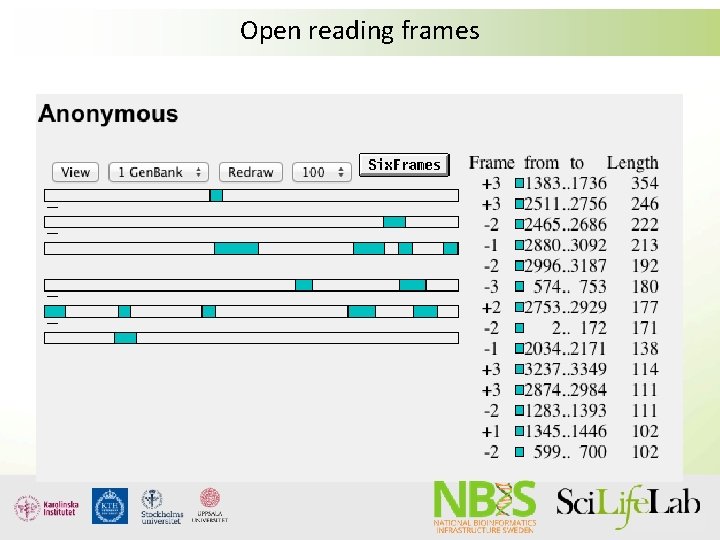
Open reading frames
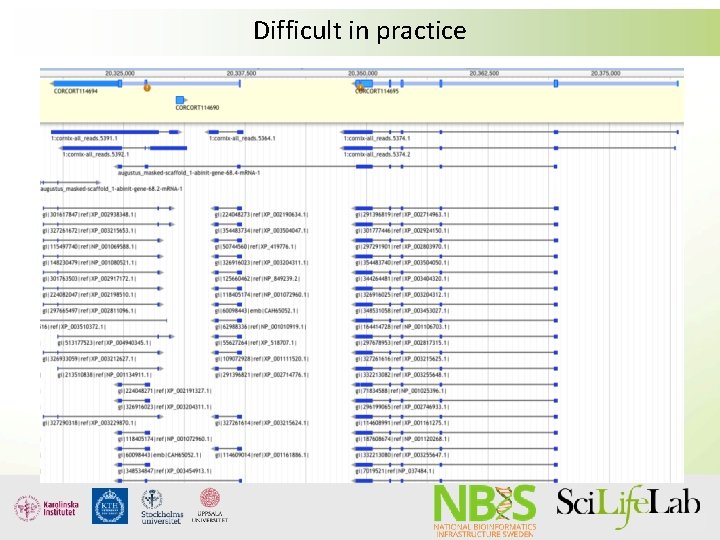
Difficult in practice
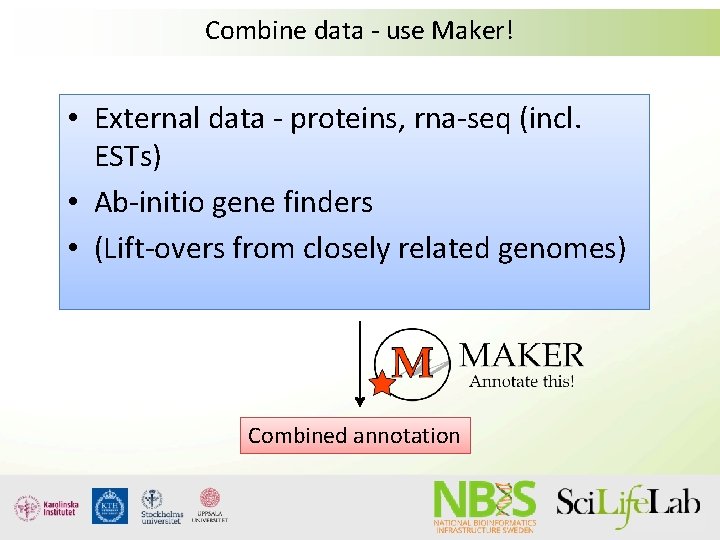
Combine data - use Maker! • External data - proteins, rna-seq (incl. ESTs) • Ab-initio gene finders • (Lift-overs from closely related genomes) Combined annotation

Transcriptomes are different but have their own challenges • No introns, but where are the start and stop codons? • Still needs functional annotation

Assembly quality • The quality of the assembly will heavily influence the quality of the annotation • SNP-errors can change start/stop-codons • Indels can cause frame-shifts • Annotation tools often have problems with incomplete loci • And of course, if a locus is completely missing from the assembly, it cannot be annotated
Assembly validation suing CEGMA/BUSCO • CEGMA now depreceted, BUSCO actively developed • Both look for core genes; CEGMA=248 core genes, BUSCO=phylogenetic groups, up to 3000 genes • Both report %complete genes -> extrapolated to amount of gene space assembled
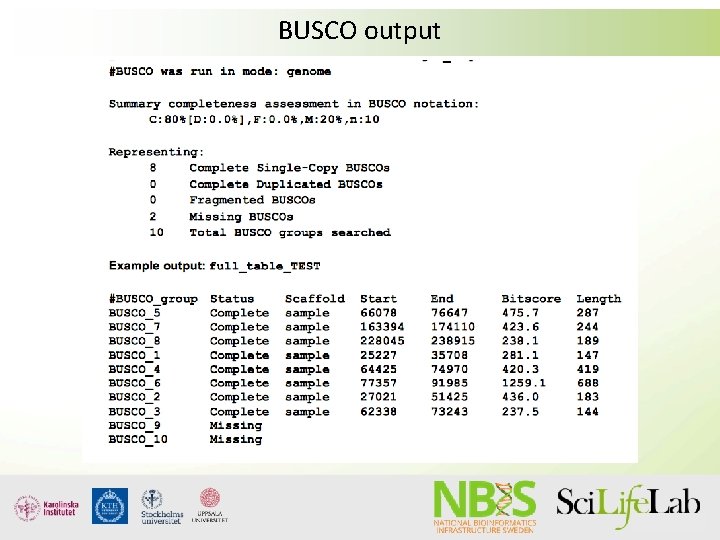
BUSCO output
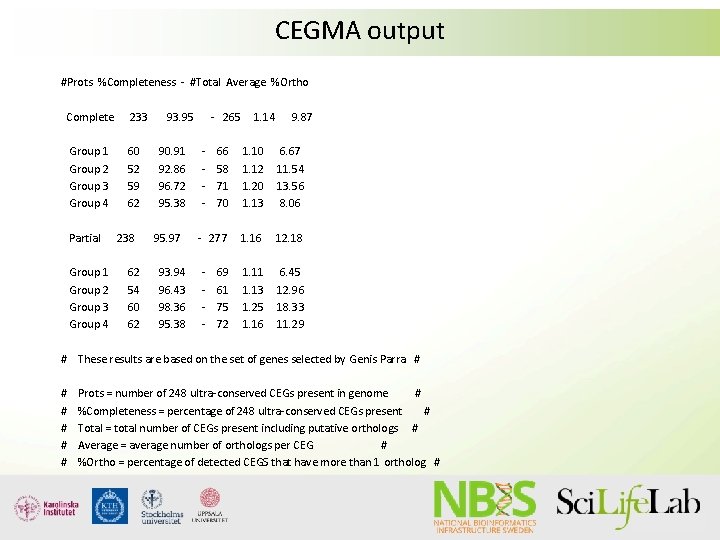
CEGMA output #Prots %Completeness - #Total Average %Ortho Complete 233 Group 1 Group 2 Group 3 Group 4 60 52 59 62 Partial Group 1 Group 2 Group 3 Group 4 238 62 54 60 62 93. 95 90. 91 92. 86 96. 72 95. 38 95. 97 93. 94 96. 43 98. 36 95. 38 - 265 - 1. 14 9. 87 66 58 71 70 1. 12 1. 20 1. 13 6. 67 11. 54 13. 56 8. 06 - 277 1. 16 12. 18 1. 11 1. 13 1. 25 1. 16 6. 45 12. 96 18. 33 11. 29 - 69 61 75 72 # These results are based on the set of genes selected by Genis Parra # # # Prots = number of 248 ultra-conserved CEGs present in genome # %Completeness = percentage of 248 ultra-conserved CEGs present # Total = total number of CEGs present including putative orthologs # Average = average number of orthologs per CEG # %Ortho = percentage of detected CEGS that have more than 1 ortholog #
Data used - Proteins
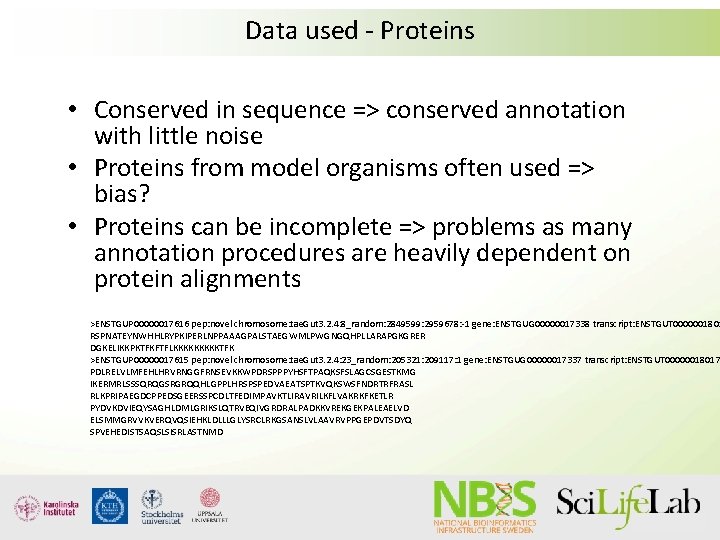
Data used - Proteins • Conserved in sequence => conserved annotation with little noise • Proteins from model organisms often used => bias? • Proteins can be incomplete => problems as many annotation procedures are heavily dependent on protein alignments >ENSTGUP 00000017616 pep: novel chromosome: tae. Gut 3. 2. 4: 8_random: 2849599: 2959678: -1 gene: ENSTGUG 00000017338 transcript: ENSTGUT 0000001801 RSPNATEYNWHHLRYPKIPERLNPPAAAGPALSTAEGWMLPWGNGQHPLLARAPGKGRER DGKELIKKPKTFKFTFLKKKKKTFK >ENSTGUP 00000017615 pep: novel chromosome: tae. Gut 3. 2. 4: 23_random: 205321: 209117: 1 gene: ENSTGUG 00000017337 transcript: ENSTGUT 00000018017 PDLRELVLMFEHLHRVRNGGFRNSEVKKWPDRSPPPYHSFTPAQKSFSLAGCSGESTKMG IKERMRLSSSQRQGSRGRQQHLGPPLHRSPSPEDVAEATSPTKVQKSWSFNDRTRFRASL RLKPRIPAEGDCPPEDSGEERSSPCDLTFEDIMPAVKTLIRAVRILKFLVAKRKFKETLR PYDVKDVIEQYSAGHLDMLGRIKSLQTRVEQIVGRDRALPADKKVREKGEKPALEAELVD ELSMMGRVVKVERQVQSIEHKLDLLLGLYSRCLRKGSANSLVLAAVRVPPGEPDVTSDYQ SPVEHEDISTSAQSLSISRLASTNMD

Data used - Proteins • Maker will align proteins for you: Blast -> Exonerate • Blast is not structure aware, Exonerate is (splice sites, start/stop codons) • Preferred file-format: fasta
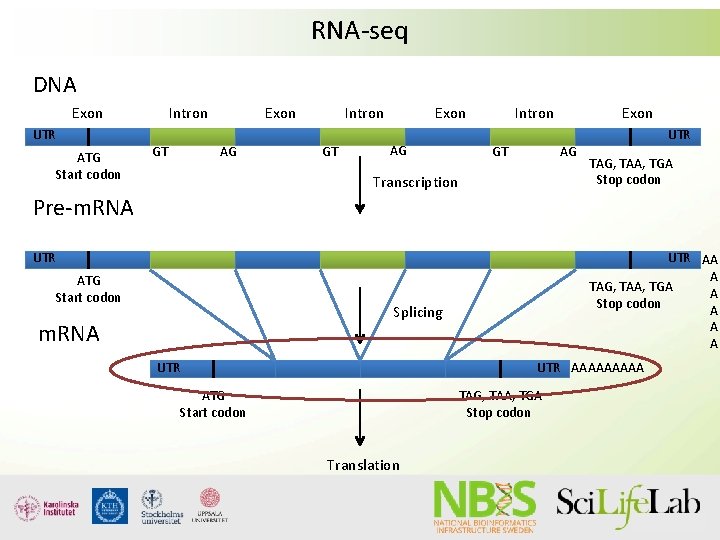
RNA-seq DNA Exon UTR ATG Start codon Intron GT Exon AG Transcription UTR TAG, TAA, TGA Stop codon Pre-m. RNA UTR ATG Start codon UTR AA A TAG, TAA, TGA A Stop codon A A A Splicing m. RNA UTR AAAAA ATG Start codon TAG, TAA, TGA Stop codon Translation
Data used - RNA-seq • Should always be included in an annotation project • From the same organism as the genomic data => unbiased • Can be very noisy (tissue/species dependent), can include pre-m. RNA • PASA, or some other filtering method, often needed
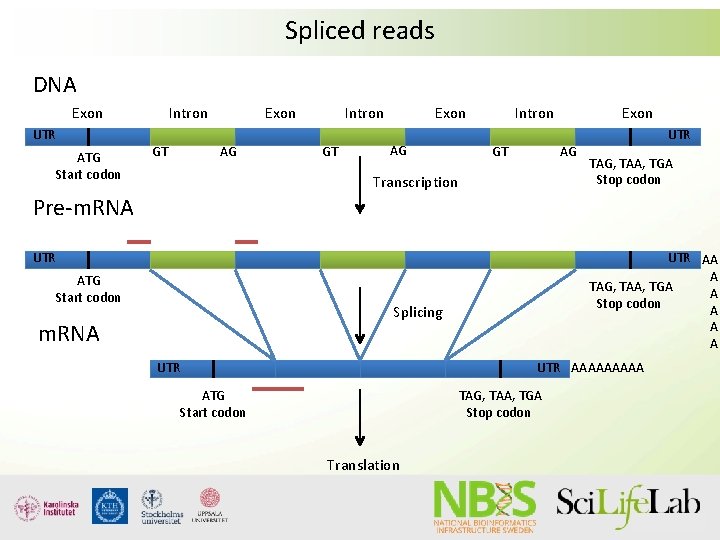
Spliced reads DNA Exon UTR ATG Start codon Intron GT Exon AG Transcription UTR TAG, TAA, TGA Stop codon Pre-m. RNA UTR ATG Start codon UTR AA A TAG, TAA, TGA A Stop codon A A A Splicing m. RNA UTR AAAAA ATG Start codon TAG, TAA, TGA Stop codon Translation

RNA-seq - Spliced reads
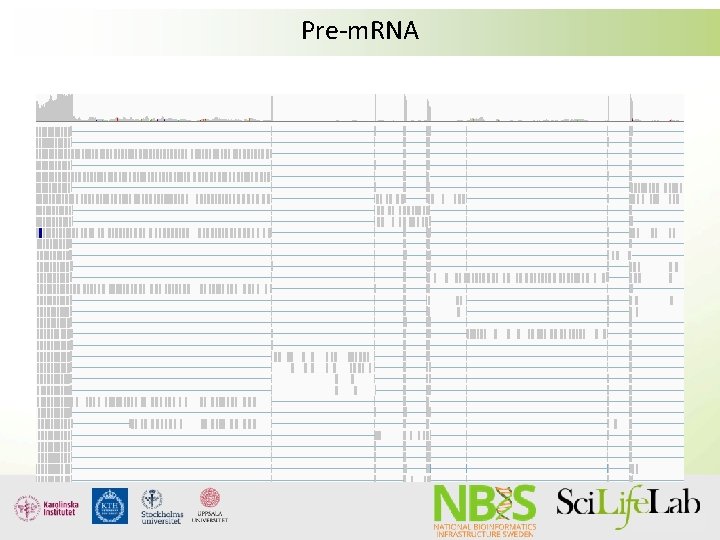
Pre-m. RNA
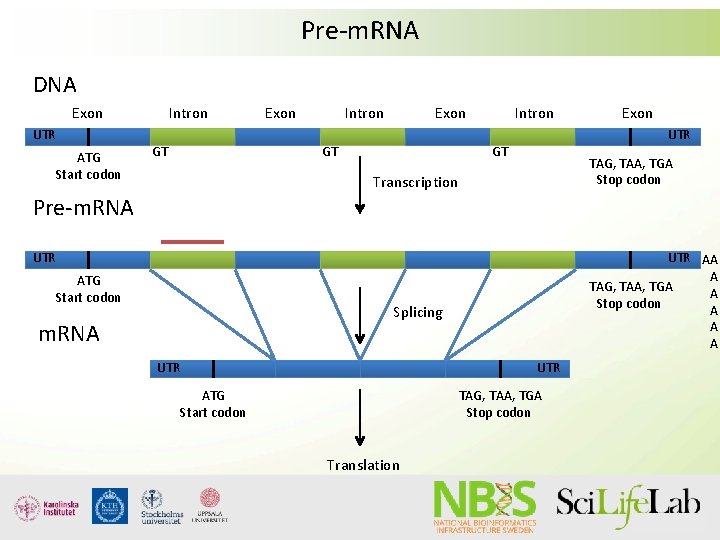
Pre-m. RNA DNA Exon UTR ATG Start codon Intron GT Exon Intron Exon GT Intron Exon UTR GT TAG, TAA, TGA Stop codon Transcription Pre-m. RNA UTR ATG Start codon UTR AA A TAG, TAA, TGA A Stop codon A A A Splicing m. RNA UTR ATG Start codon TAG, TAA, TGA Stop codon Translation
Pre-m. RNA
A lot is transcribed in a cell
Stranded rna-seq
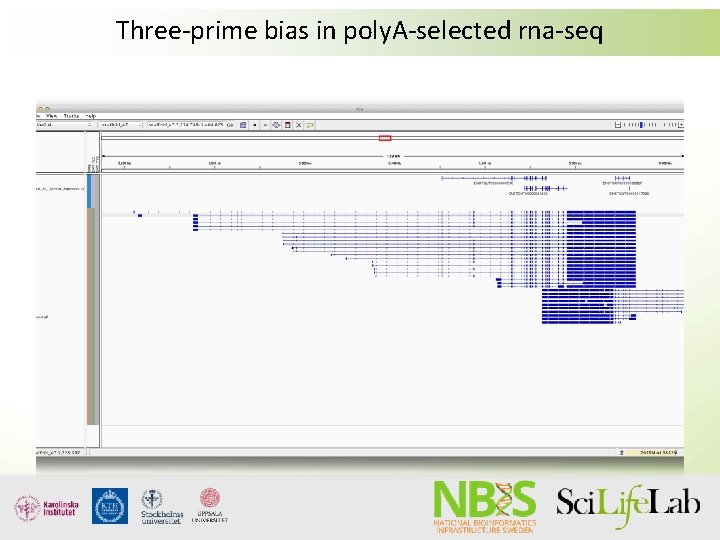
Three-prime bias in poly. A-selected rna-seq
How to use RNA-seq • Maker will align transcripts (ESTs), but these need to be assembled first. • Cufflinks: mapped reads -> transcripts
How to use RNA-seq • Maker will align transcripts (ESTs), but these need to be assembled first. • Cufflinks: mapped reads -> transcripts • Trinity: assembles transcripts without a genome
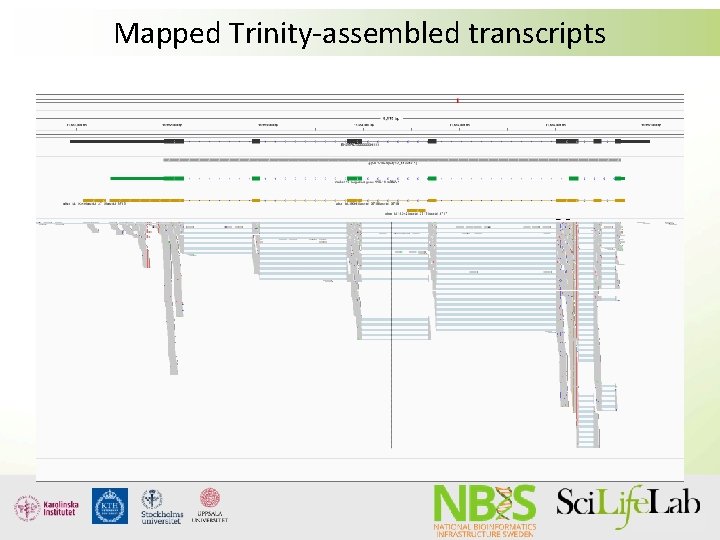
Mapped Trinity-assembled transcripts
How to use RNA-seq • Maker will align transcripts (ESTs), but these need to be assembled first. • Cufflinks: mapped reads -> transcripts • Trinity: assembles transcripts without a genome • PASA can be used to improve transcript quality
Ab initio gene finders are used in Maker • Commonly used programs: Augustus, Snap, Genemark-ES, FGENESH, Genscan, Glimmer. HMM, … • Uses HMM-models to figure out how introns, exons, UTRs etc. are structured • These HMM-models need to be trained!
Liftovers are very useful for orthology determination • Kraken • Align the two genomes (Satsuma) and then transfer annotations between aligned regions

General recommendations • Always combine different types of evidence! • One single method is not enough! • Use Maker!

Transcript annotation • Here the transcript is already defined. The challenge is to find where the coding regions starts and stops • Transdecoder
Transdecoder
Transdecoder

Or get help - NBIS assembly and annotation team • Five people working with assembly and annotation • Deliver high quality annotations • Enable visualization and manual curation through a web interface • Also available for consultation • http: //nbis. se/supportform/index. ph p

Biosupport. se
- Slides: 47