Introduction to Cell biology Hierarchical organisation of the

Introduction to Cell biology
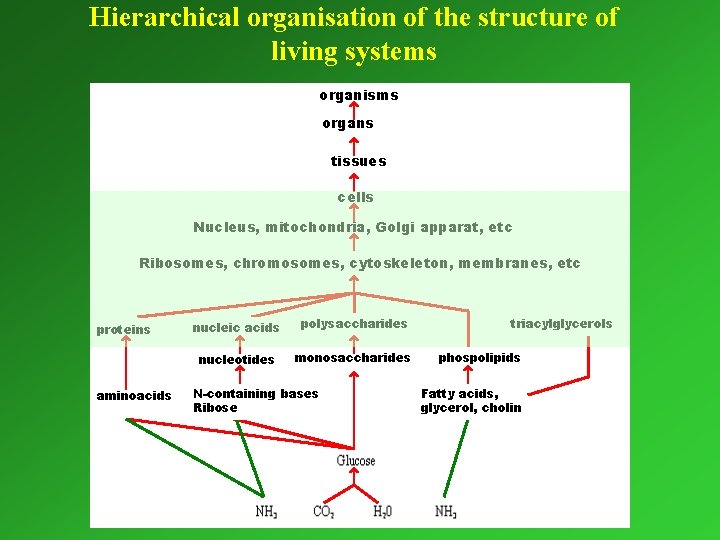
Hierarchical organisation of the structure of living systems organisms organs tissues cells Nucleus, mitochondria, Golgi apparat, etc Ribosomes, chromosomes, cytoskeleton, membranes, etc proteins aminoacids nucleic acids polysaccharides nucleotides monosaccharides N-containing bases Ribose triacylglycerols phospolipids Fatty acids, glycerol, cholin
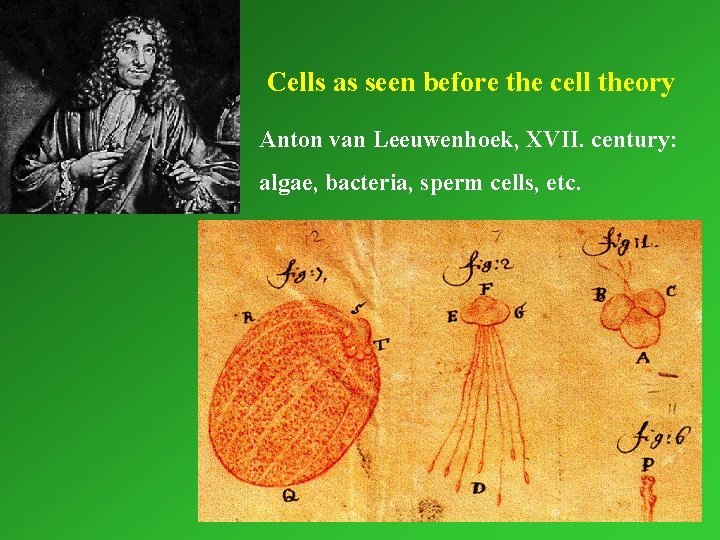
Cells as seen before the cell theory Anton van Leeuwenhoek, XVII. century: algae, bacteria, sperm cells, etc.

Robert Hooke 1665: „cell”: unit in dead samples of cork.

The cell theory Cell as the central unit of biological organization • Cells are the basic units of life. • All living organisms are made up of cells. • Only living cells can produce new cells. Matthias Schleiden 1838 Theodor Schwann 1835 plants are made up of cells animals are made up of cells

Rudolf Virchow 1858: „Every animal appears as the sum of vital units, each of which bears in itself the complete characteristics of life”

Louis Pasteur 1865 : „Spontaneous generation” of life ruled out experimentally „There is now no circumstance known in which it can be affirmed that microscopic beings came into the world without germs, without parents similar to themselves. "

Tranzitions from non-living towards living: I. Prions: molecules resembling ion channels, causing serious illnesses
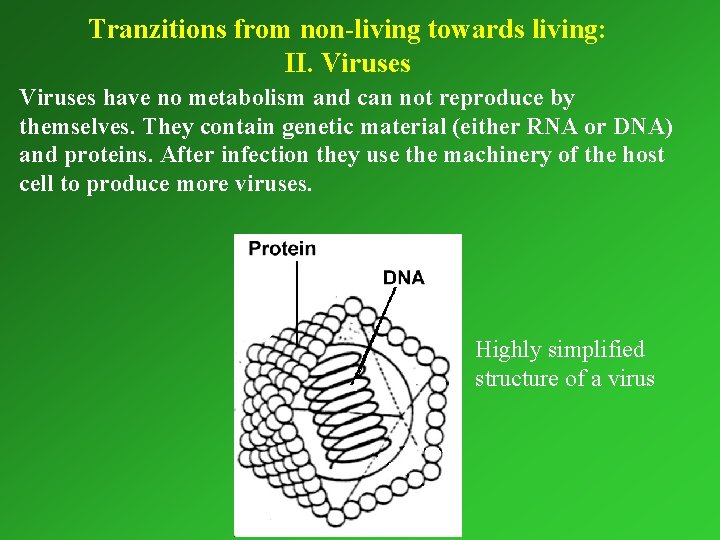
Tranzitions from non-living towards living: II. Viruses have no metabolism and can not reproduce by themselves. They contain genetic material (either RNA or DNA) and proteins. After infection they use the machinery of the host cell to produce more viruses. Highly simplified structure of a virus
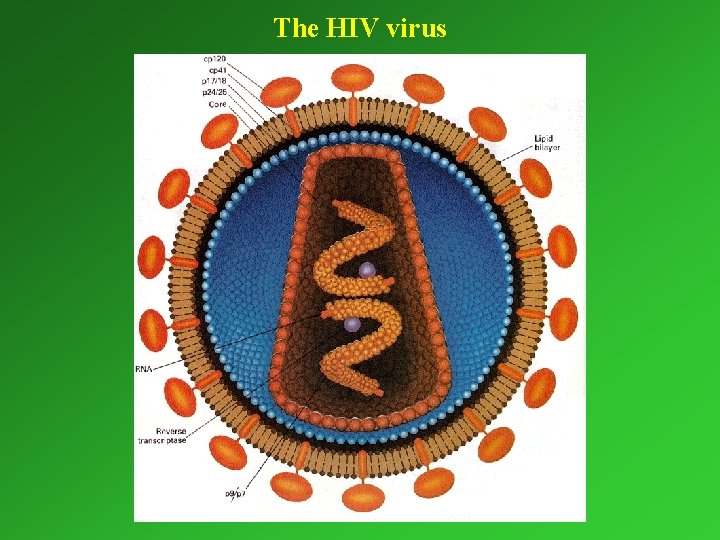
The HIV virus
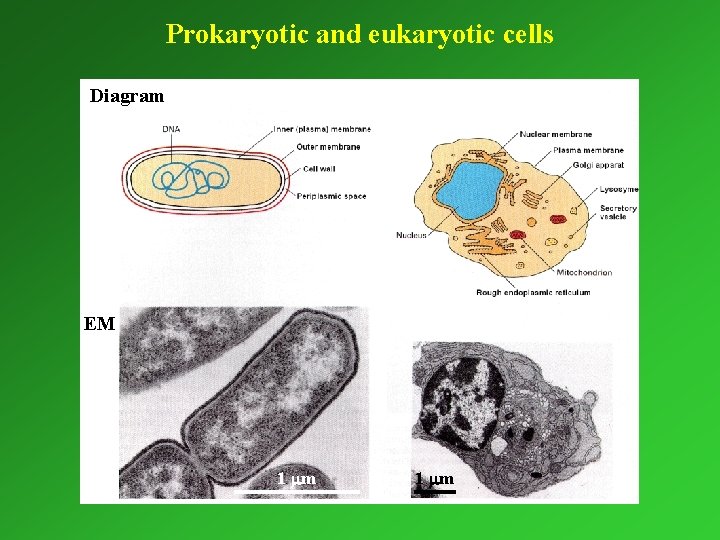
Prokaryotic and eukaryotic cells Diagram EM 1 mm

I. (BIO)CHEMICAL FOUNDATIONS

The most important groups of organic molecules: Proteins composed of amino acids Lipids composed of glycerol and fatty acids Carbohydrates: mono-, oligo- and polysaccharides Nucleic acids: DNA, RNAs

I. /1. PROTEINS Classification of proteins: • Enzymes • Receptors • Transport proteins • Storage proteins (casein in milk, ferritin /iron/) • Contractile proteins • Structural proteins • Immune proteins • Regulatory proteins • Others (e. g. antifreeze proteins)
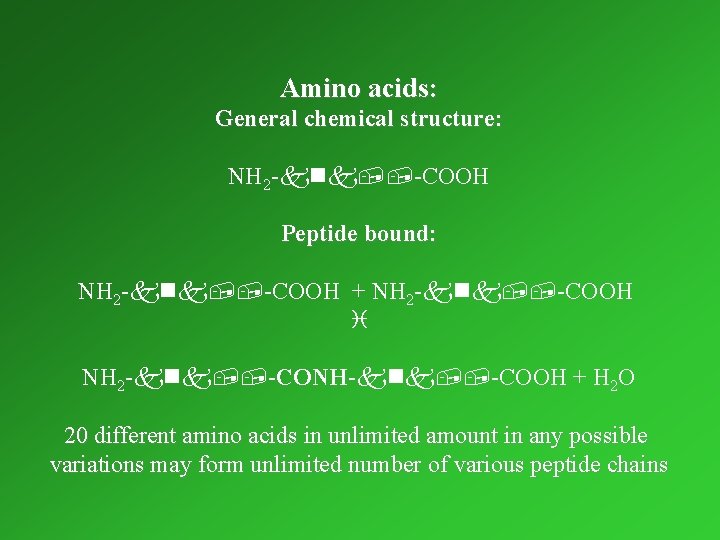
Amino acids: General chemical structure: NH 2 -knk, , -COOH Peptide bound: NH 2 -knk, , -COOH + NH 2 -knk, , -COOH i NH 2 -knk, , -CONH-knk, , -COOH + H 2 O 20 different amino acids in unlimited amount in any possible variations may form unlimited number of various peptide chains
Primary structure or sequence: linear arrangement of the amino acids that constitute the polypeptide chain Sequencing: to determine the order of amino acids of a protein. Sequence motive: Examples: a specific amino acid arrangement that appears in several different proteins and play the same role in these proteins. DNA binding motive signal sequence (transport of the protein to a given organelle) sequence for phosphorylation ligand-binding sequences (e. g. ATP, growth hormons)
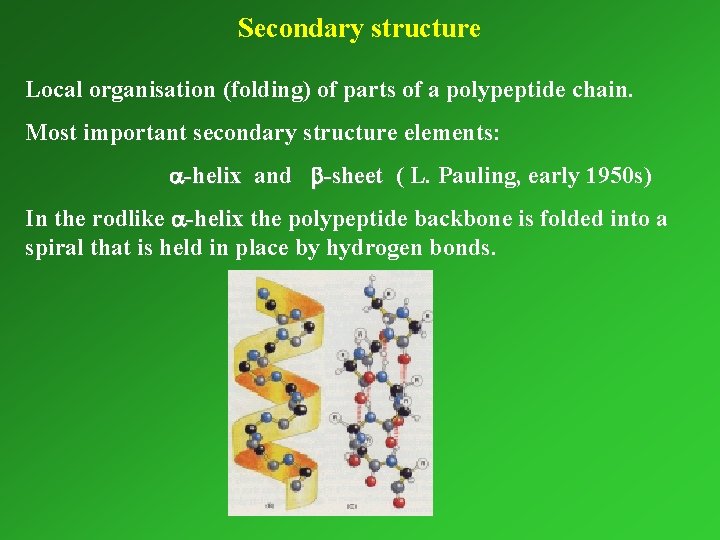
Secondary structure Local organisation (folding) of parts of a polypeptide chain. Most important secondary structure elements: a-helix and b-sheet ( L. Pauling, early 1950 s) In the rodlike a-helix the polypeptide backbone is folded into a spiral that is held in place by hydrogen bonds.
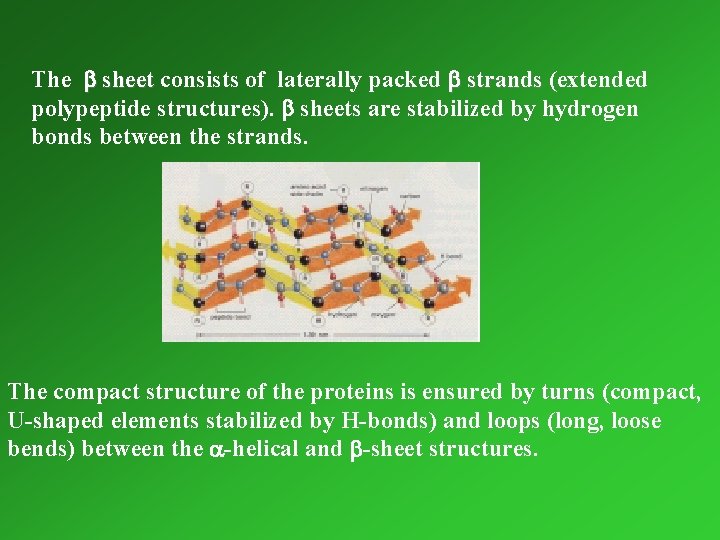
The b sheet consists of laterally packed b strands (extended polypeptide structures). b sheets are stabilized by hydrogen bonds between the strands. The compact structure of the proteins is ensured by turns (compact, U-shaped elements stabilized by H-bonds) and loops (long, loose bends) between the a-helical and b-sheet structures.
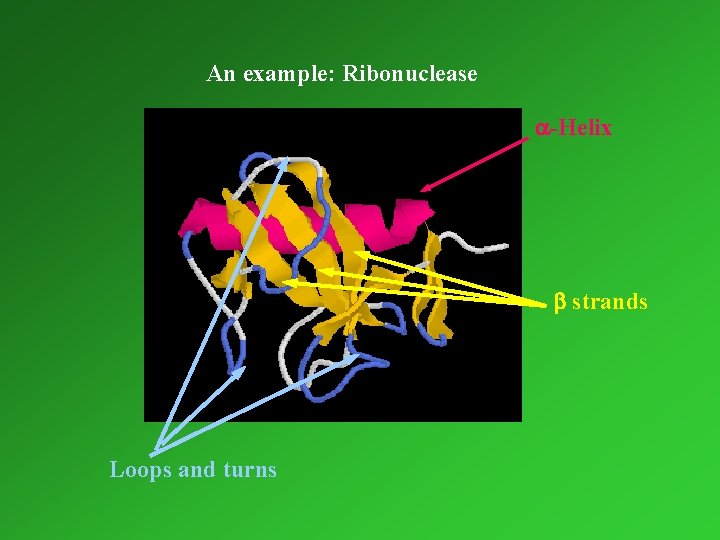
An example: Ribonuclease a-Helix b strands Loops and turns

Tertiary and quaternary structure Tertiary structure: Three-dimensional arrangement of all amino acids, which results in mainly from hydrophobic interactions between nonpolar amino acid side-chains. These interactions hold helices, strands and coils together. The highest level of organisation for monomeric proteins. Quaternary structure: structure number and relative positions of subunits in multimeric proteins. Determination of the three-dimensional structure of proteins: x-ray crystallography nuclear magnetic resonance (NMR)
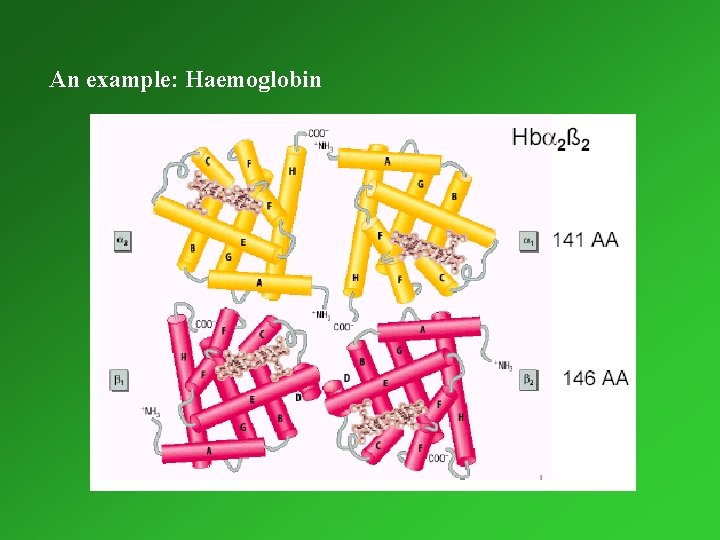
An example: Haemoglobin
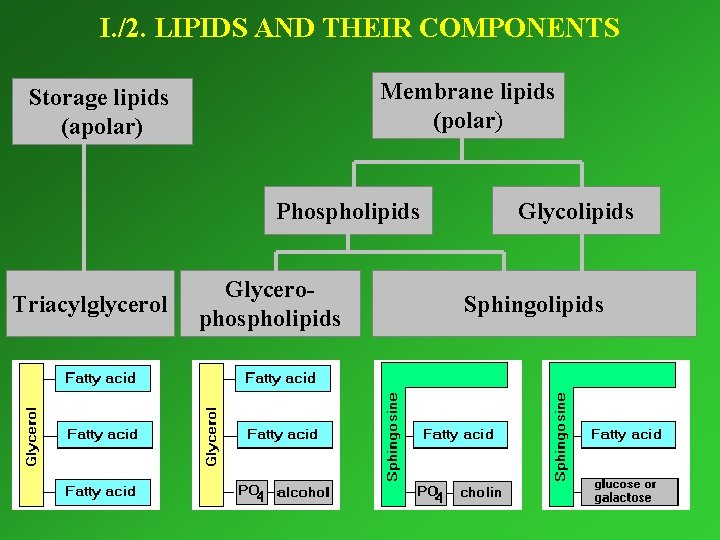
I. /2. LIPIDS AND THEIR COMPONENTS Membrane lipids (polar) Storage lipids (apolar) Phospholipids Triacylglycerol Glycerophospholipids Glycolipids Sphingolipids
Triacylglycerols Serve for storage (lipid droplets in fat cells) and isolation.
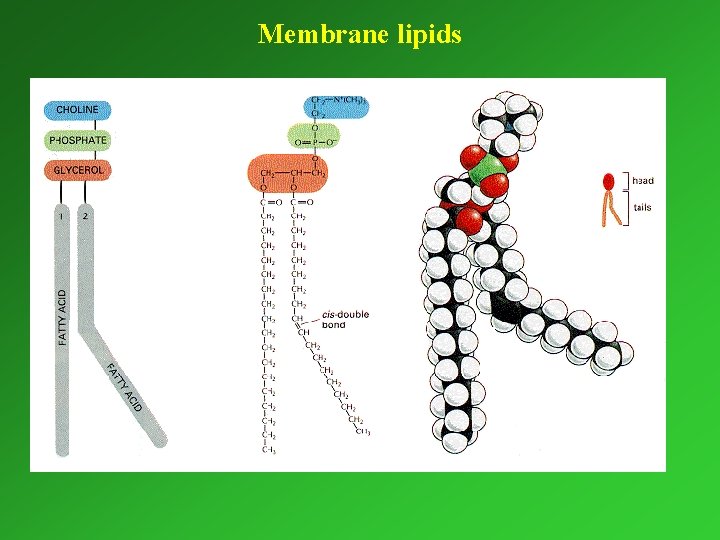
Membrane lipids
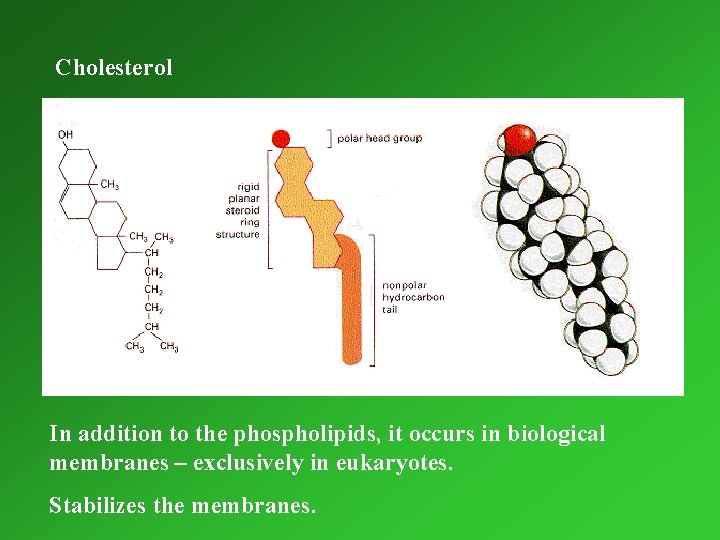
Cholesterol In addition to the phospholipids, it occurs in biological membranes – exclusively in eukaryotes. Stabilizes the membranes.

I. /3. CARBOHYDRATES The most abundant biomolecules on the earth. Essential components of foodstuff (sugar) Forms of occurence in living systems: monosaccharides (e. g. glucose) oligosaccharides (e. g. saccharose, lactose) polysaccharides (e. g. glycogene, starch) Occurrence in complex macromolecules: with lipids (e. g. glycolipides) with proteins (glycoproteins and proteoglycans) within nucleic acids (constituents of RNA and DNA)
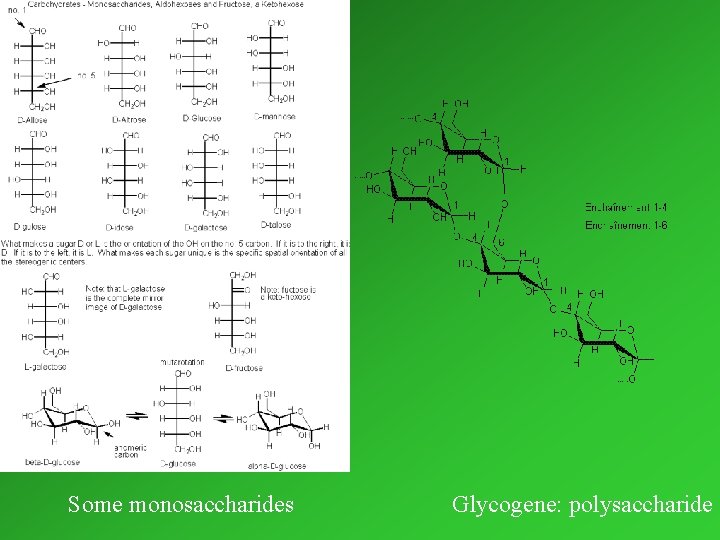
Some monosaccharides Glycogene: polysaccharide
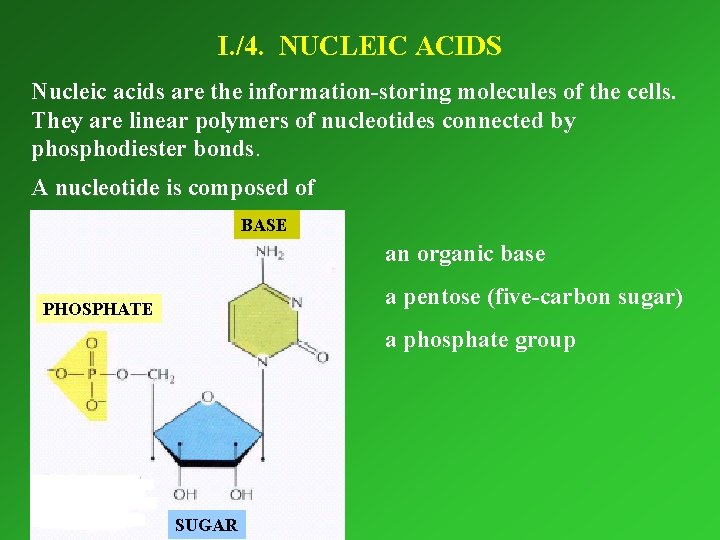
I. /4. NUCLEIC ACIDS Nucleic acids are the information-storing molecules of the cells. They are linear polymers of nucleotides connected by phosphodiester bonds. A nucleotide is composed of BASE an organic base a pentose (five-carbon sugar) PHOSPHATE a phosphate group SUGAR
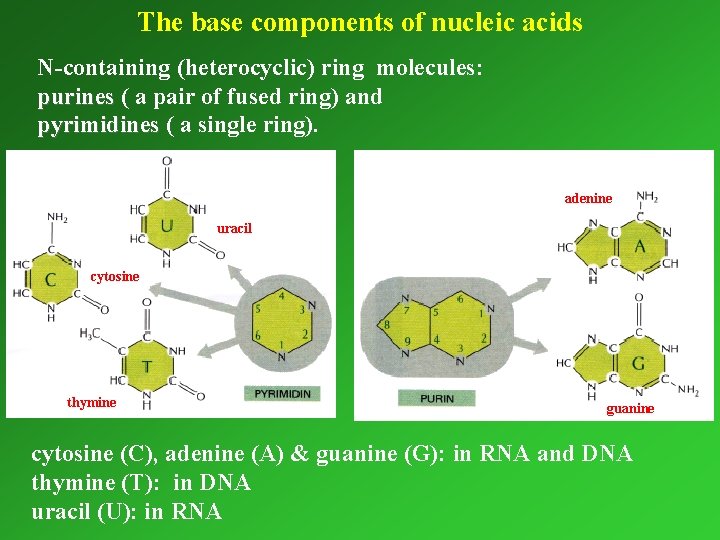
The base components of nucleic acids N-containing (heterocyclic) ring molecules: purines ( a pair of fused ring) and pyrimidines ( a single ring). adenine uracil cytosine thymine guanine cytosine (C), adenine (A) & guanine (G): in RNA and DNA thymine (T): in DNA uracil (U): in RNA
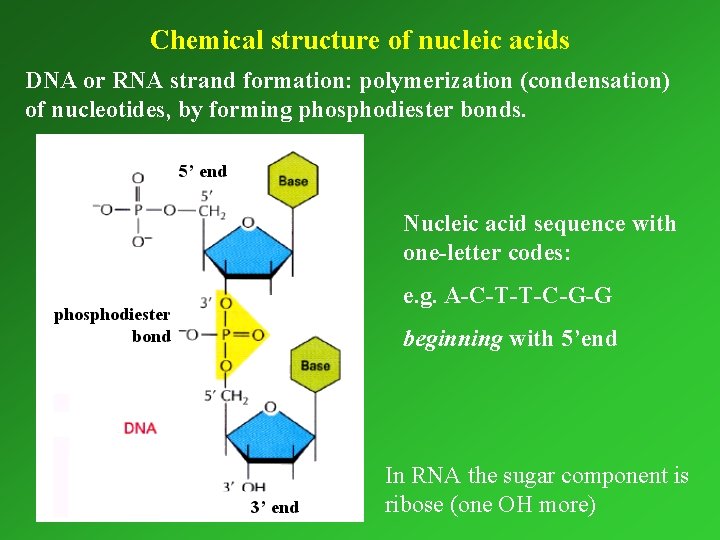
Chemical structure of nucleic acids DNA or RNA strand formation: polymerization (condensation) of nucleotides, by forming phosphodiester bonds. Nucleic acid sequence with one-letter codes: e. g. A-C-T-T-C-G-G beginning with 5’end In RNA the sugar component is ribose (one OH more)
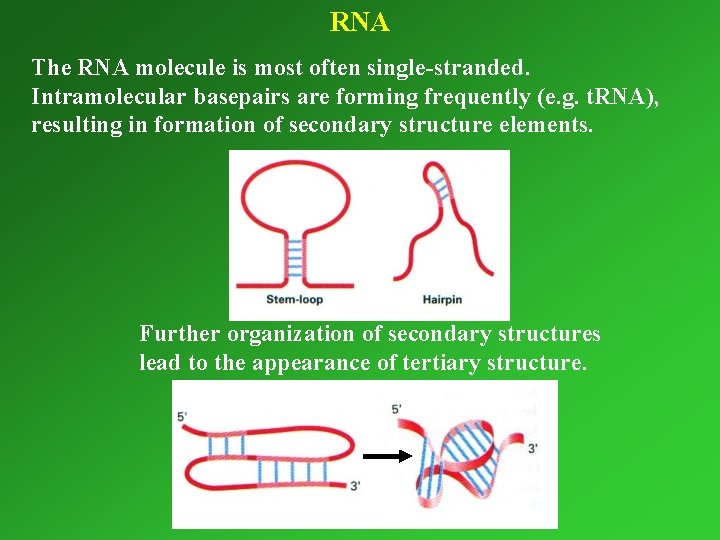
RNA The RNA molecule is most often single-stranded. Intramolecular basepairs are forming frequently (e. g. t. RNA), resulting in formation of secondary structure elements. Further organization of secondary structures lead to the appearance of tertiary structure.

• A considerable fraction of RNA occurs in great complexes together with proteins (e. g. ribosomes) • RNA can have catalytic activity (ribozymes). • RNA is the genetic material in several viruses (polio, influenza, rota, HIV, etc).
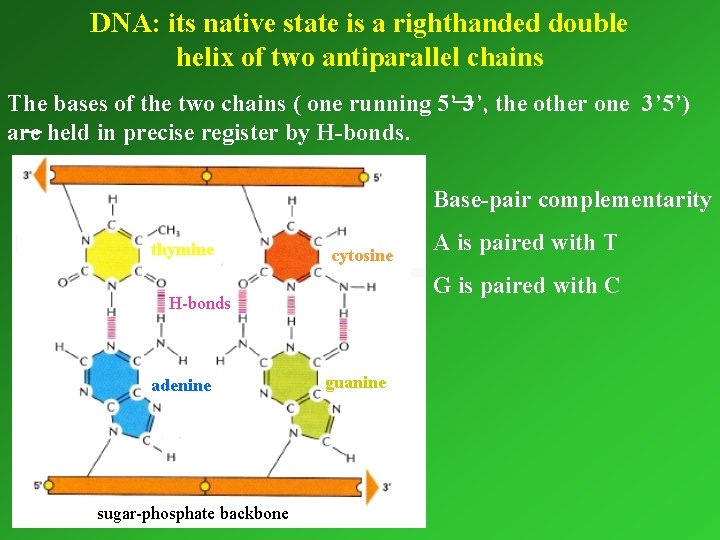
DNA: its native state is a righthanded double helix of two antiparallel chains The bases of the two chains ( one running 5’ 3’, the other one 3’ 5’) are held in precise register by H-bonds. Base-pair complementarity thymine cytosine G is paired with C H-bonds adenine sugar-phosphate backbone A is paired with T guanine
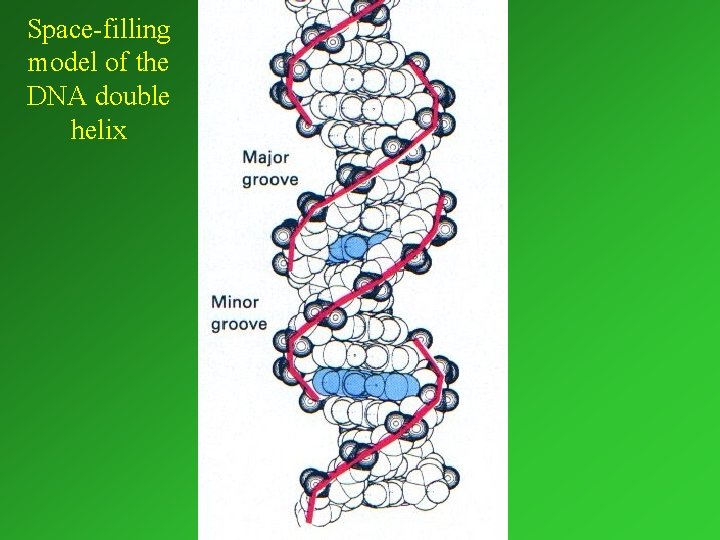
Space-filling model of the DNA double helix

Nobel Prize 1962 „for their discoveries concerning the molecular structure of nucleic acids and its significance for information transfer in living material” James Dewey Watson Francis Harry Compton Crick Harvard University Cambridge, MA, USA Institute of Molecular Biology Cambridge, United Kingdom
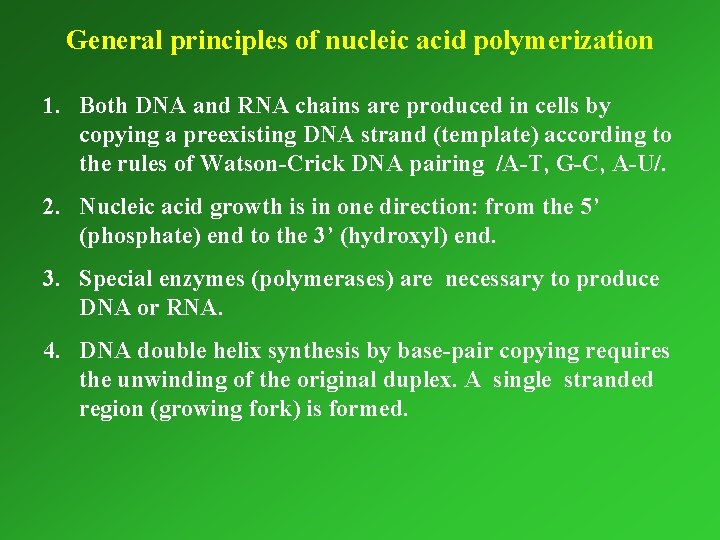
General principles of nucleic acid polymerization 1. Both DNA and RNA chains are produced in cells by copying a preexisting DNA strand (template) according to the rules of Watson-Crick DNA pairing /A-T, G-C, A-U/. 2. Nucleic acid growth is in one direction: from the 5’ (phosphate) end to the 3’ (hydroxyl) end. 3. Special enzymes (polymerases) are necessary to produce DNA or RNA. 4. DNA double helix synthesis by base-pair copying requires the unwinding of the original duplex. A single stranded region (growing fork) is formed.
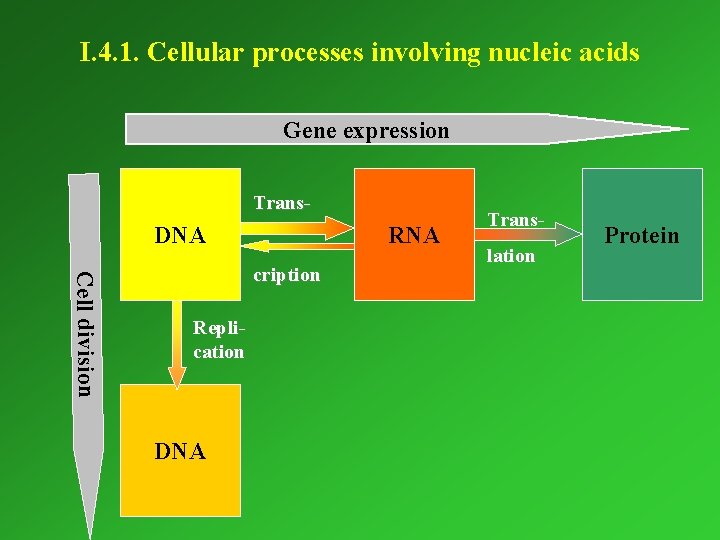
I. 4. 1. Cellular processes involving nucleic acids Gene expression Trans- DNA RNA Cell division cription Replication DNA Translation Protein
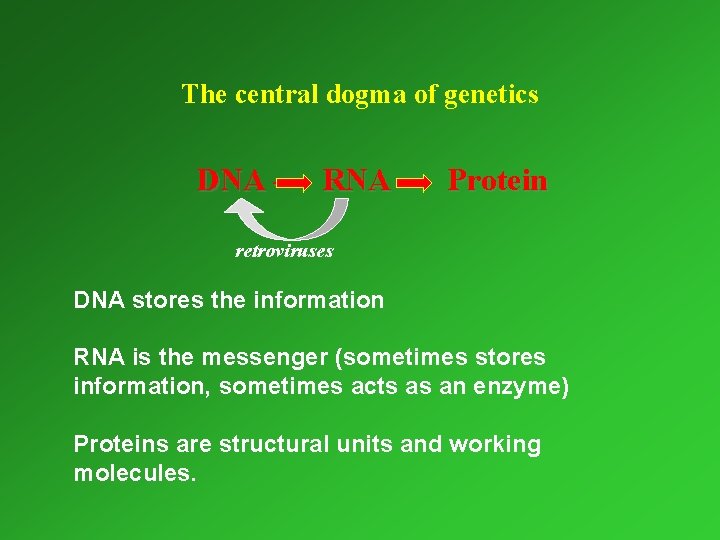
The central dogma of genetics DNA – RNA – Protein retroviruses DNA stores the information RNA is the messenger (sometimes stores information, sometimes acts as an enzyme) Proteins are structural units and working molecules.
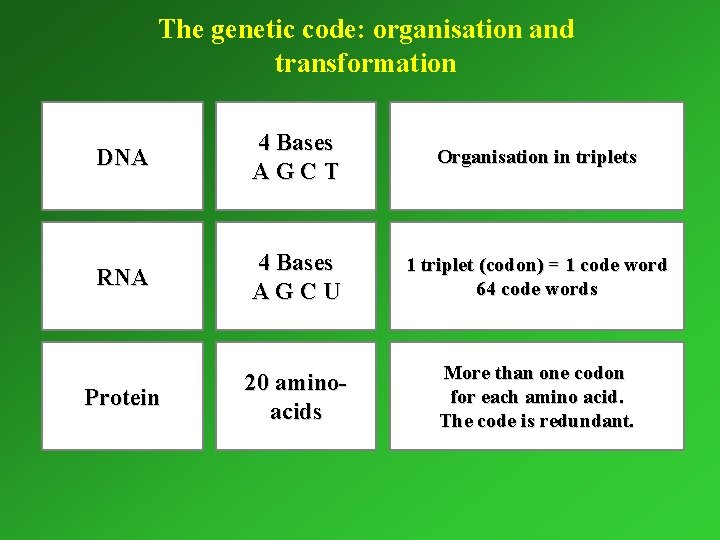
The genetic code: organisation and transformation DNA 4 Bases AGCT Organisation in triplets RNA 4 Bases AGCU 1 triplet (codon) = 1 code word 64 code words Protein 20 aminoacids More than one codon for each amino acid. The code is redundant.
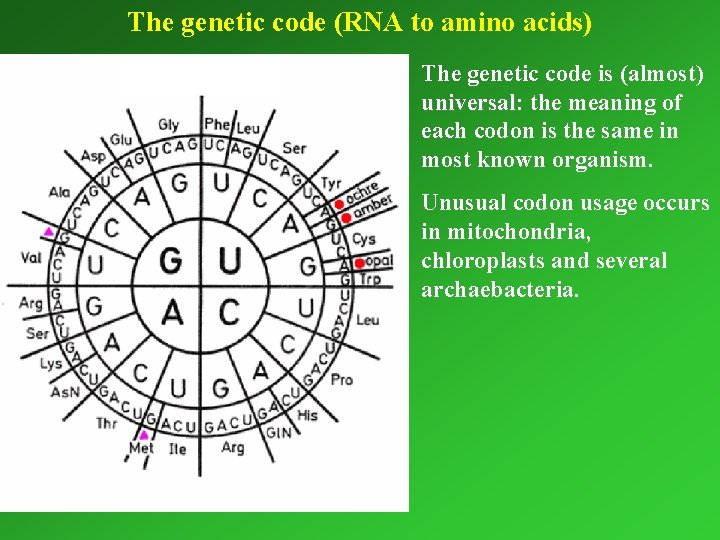
The genetic code (RNA to amino acids) The genetic code is (almost) universal: universal the meaning of each codon is the same in most known organism. Unusual codon usage occurs in mitochondria, chloroplasts and several archaebacteria.

Reading frames The genetic code is commaless! Thus: 5’___ GCUUGUUAACGAAUUA__ __GCUUGUUAACGAAUUA Ala--Cys--Leu--Arg--Ile m. RNA __GCUUGUUAACGAAUUA Leu--Val--Tyr--Glu--Leu

I. 4. 2. Gene and genome Gene: Gene The nucleotide sequence needed to produce a functionally competent „working molecule” (RNA or protein Genome: The totality of the genes of a given organism.

Genome Sequence Projects Since 1995 the following complete genom sequences became available: Prokaryotes: More than 30 Bakterial species (several disease-causing ones), some Archaebakteria Eukaryotes: Saccharomyces cerevisae (baker’s yeast) Caenorhabditis elegans (worm) Drosophila melanogaster (fruitfly) Arabidopsis thaliana (plant) Mus musculus (mouse) Homo sapiens


The human genome • the sequence of the human genom contains 3, 3 billion bases, organised in 24 chromosomes (22, X, Y) • 30 000 to 40 000 genes • 233 genes are evidently of bacterial origin • 98 % of the sequence is „nonfunctional” • the genetic identity of the human beings is 99. 9 % Nature, 15. February 2001/Science, 16. February 2001
1. 4. 3. Gene expression: expression the entire cellular process whereby the information encoded in a particular gene is decoded to a particular protein. Molecular processes involved in gene expression: transcription und translation. During transcription an RNA (messenger RNA, m. RNA) is synthesized, which contains the genetic information of the DNA as a complementary sequence. The procedure is catalyzed by DNA dependent RNA polymerases. During translation the nucleotide sequence of the m. RNA is converted to amino acid sequence of a protein. Besides the m. RNA, ribosomes and t. RNAs numerous enzymes and regulator proteins play important roles in this procedure.
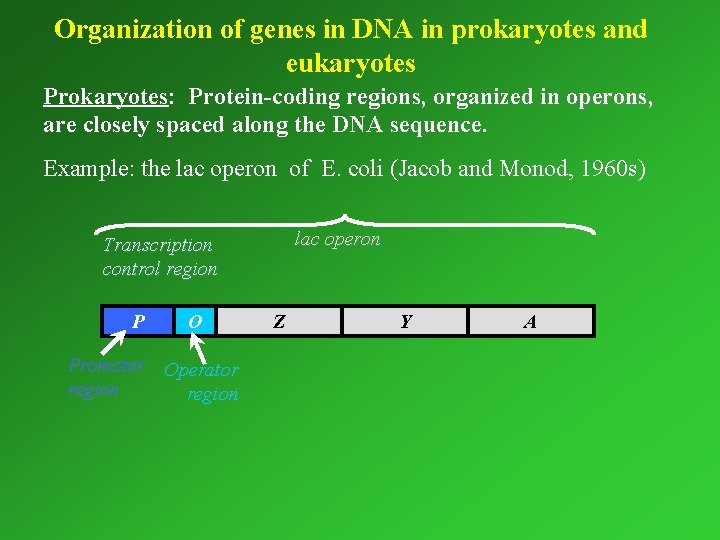
Organization of genes in DNA in prokaryotes and eukaryotes Prokaryotes: Protein-coding regions, organized in operons, operons are closely spaced along the DNA sequence. Example: the lac operon of E. coli (Jacob and Monod, 1960 s) lac operon Transcription control region P O Promoter Operator region Z Y A

Eukaryotes: a considerable amount of DNA is untranslated Transcribed regions of most of the genes is composed of several exons (translated from m. RNA) and introns (eliminated from m. RNA before translation). Example: human beta globin gene: 50 90 130 222 Untranslated regions Exons Introns 850 126 132
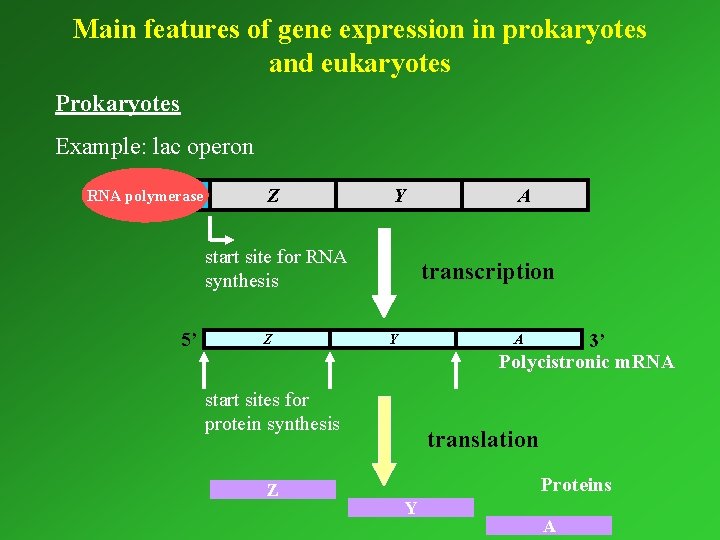
Main features of gene expression in prokaryotes and eukaryotes Prokaryotes Example: lac operon RNA polymerase P O Z Y start site for RNA synthesis 5’ Z transcription 3’ Polycistronic m. RNA Y A start sites for protein synthesis Z A translation Proteins Y A
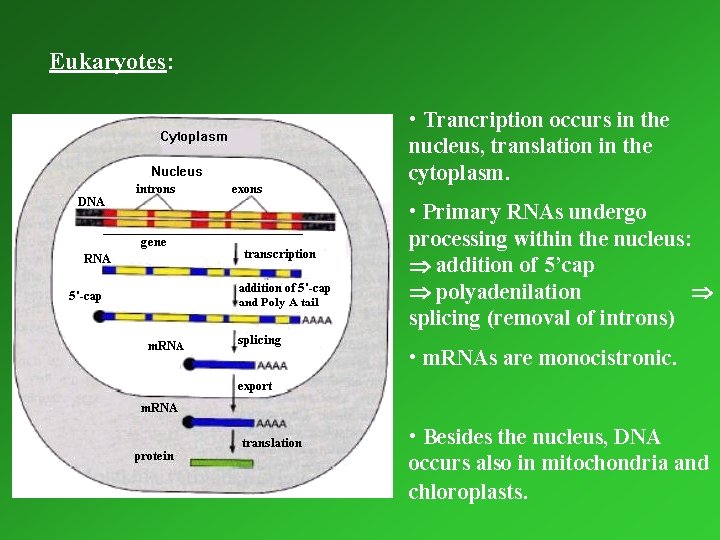
Eukaryotes: • Trancription occurs in the nucleus, translation in the cytoplasm. • Primary RNAs undergo processing within the nucleus: addition of 5’cap polyadenilation splicing (removal of introns) • m. RNAs are monocistronic. • Besides the nucleus, DNA occurs also in mitochondria and chloroplasts.
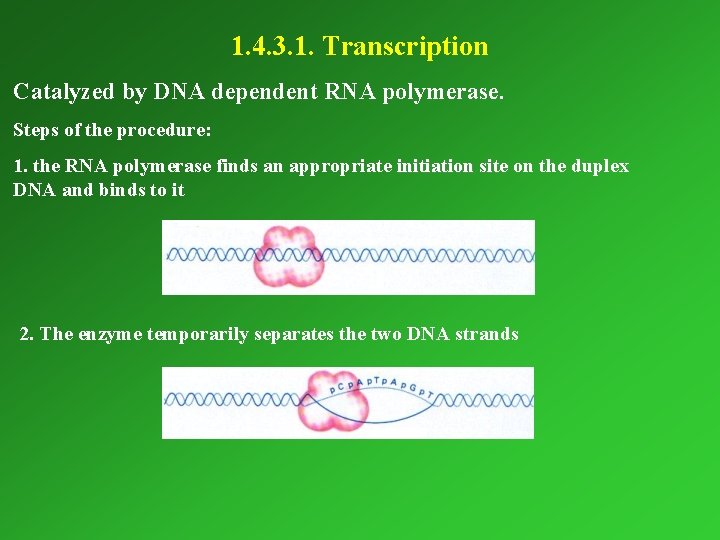
1. 4. 3. 1. Transcription Catalyzed by DNA dependent RNA polymerase. Steps of the procedure: 1. the RNA polymerase finds an appropriate initiation site on the duplex DNA and binds to it 2. The enzyme temporarily separates the two DNA strands
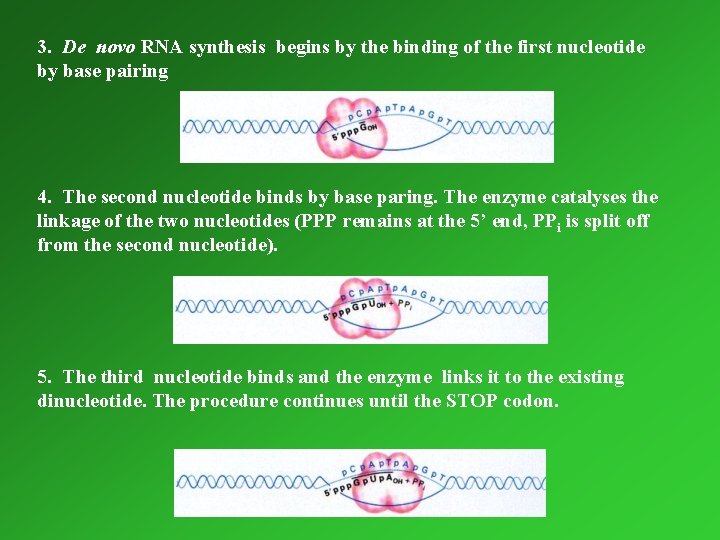
3. De novo RNA synthesis begins by the binding of the first nucleotide by base pairing 4. The second nucleotide binds by base paring. The enzyme catalyses the linkage of the two nucleotides (PPP remains at the 5’ end, PPi is split off from the second nucleotide). 5. The third nucleotide binds and the enzyme links it to the existing dinucleotide. The procedure continues until the STOP codon.
1. 4. 3. 2. Translation. Participants: • m. RNA: m. RNA source of the genetic information • loaded t. RNA: adaptormolecule, recognizing the codon and providing the corresponding amino acid. • Ribosomes: Ribosomes the „machines” in which the proteins are produced on the basis of the genetic information provided by m. RNA. • Numerous other proteins serving as regulators: intiationelongation and terminationfactors • GTP and ATP
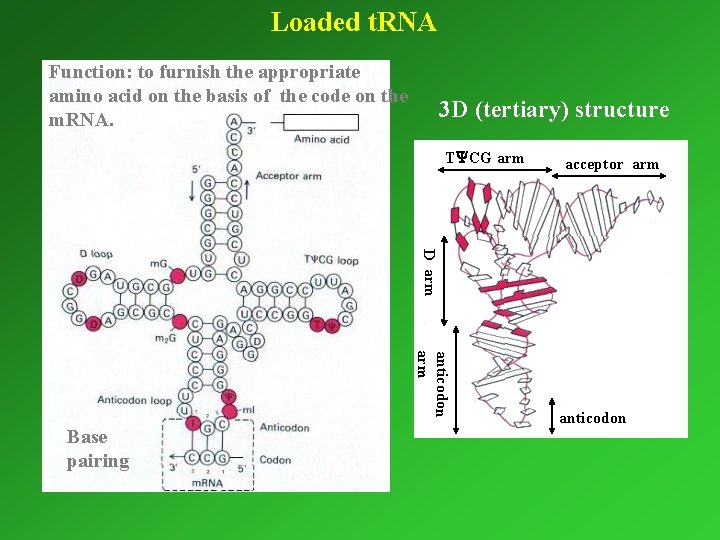
Loaded t. RNA Function: to furnish the appropriate amino acid on the basis of the code on the m. RNA. 3 D (tertiary) structure T CG arm acceptor arm D arm anticodon arm Base pairing anticodon
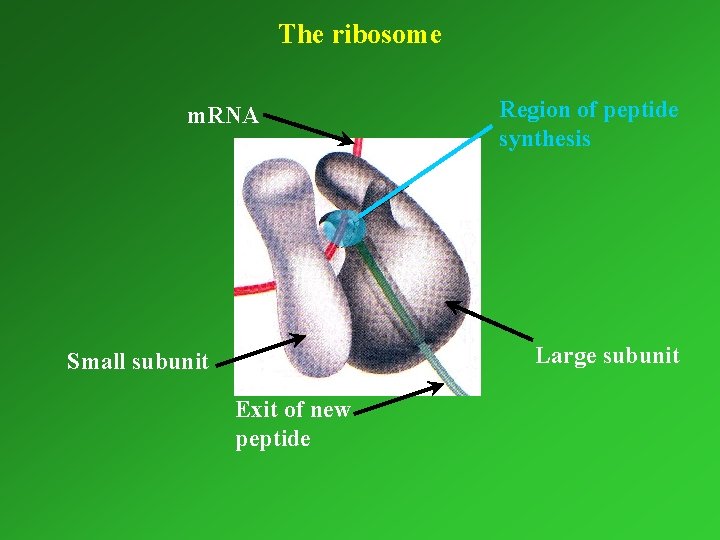
The ribosome m. RNA Region of peptide synthesis Large subunit Small subunit Exit of new peptide
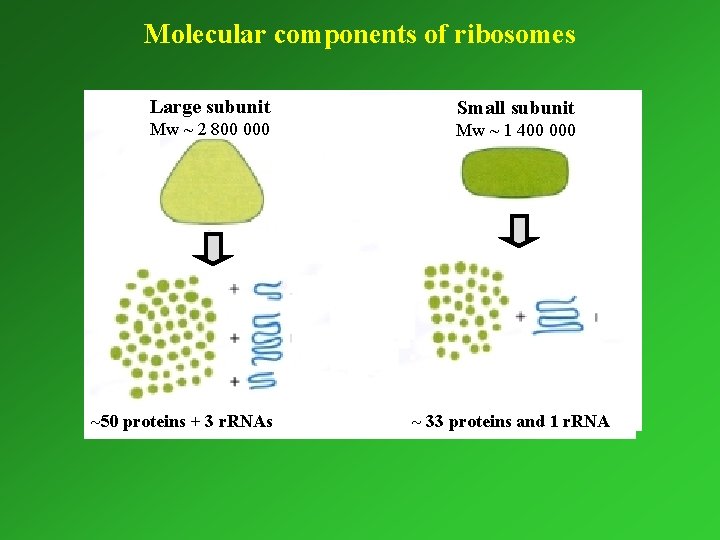
Molecular components of ribosomes Large subunit Small subunit Mw ~ 2 800 000 Mw ~ 1 400 000 ~50 proteins + 3 r. RNAs ~ 33 proteins and 1 r. RNA
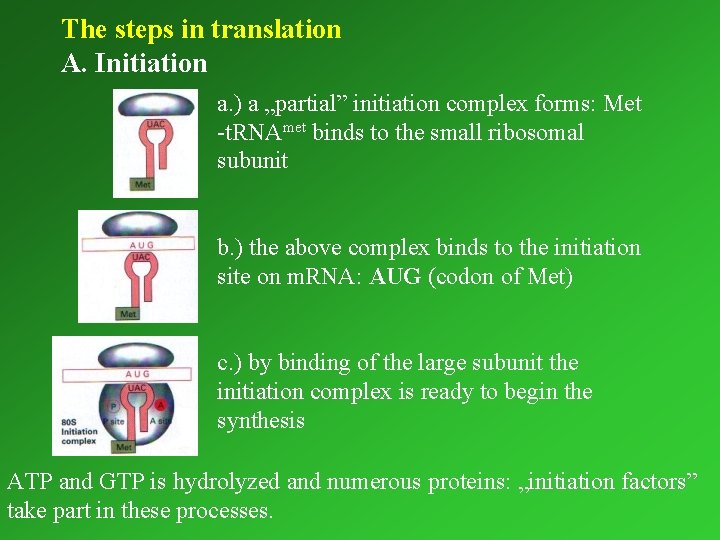
The steps in translation A. Initiation a. ) a „partial” initiation complex forms: Met -t. RNAmet binds to the small ribosomal subunit b. ) the above complex binds to the initiation site on m. RNA: AUG (codon of Met) c. ) by binding of the large subunit the initiation complex is ready to begin the synthesis ATP and GTP is hydrolyzed and numerous proteins: „initiation factors” take part in these processes.
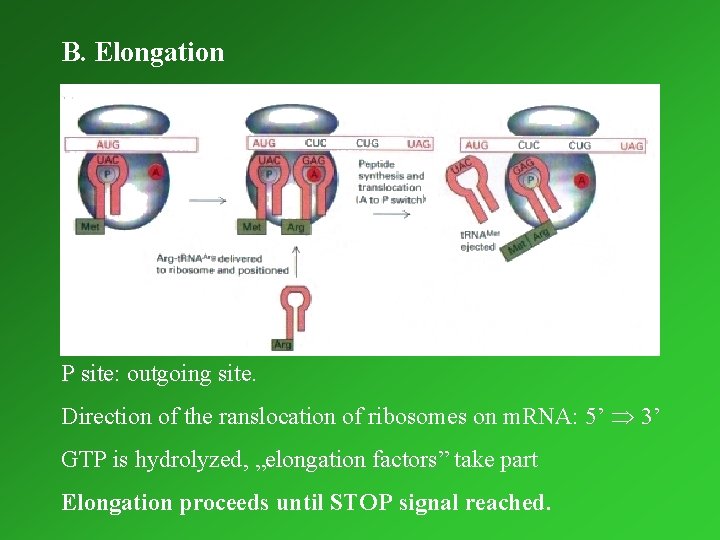
B. Elongation P site: site outgoing site. Direction of the ranslocation of ribosomes on m. RNA: 5’ 3’ GTP is hydrolyzed, „elongation factors” take part Elongation proceeds until STOP signal reached.
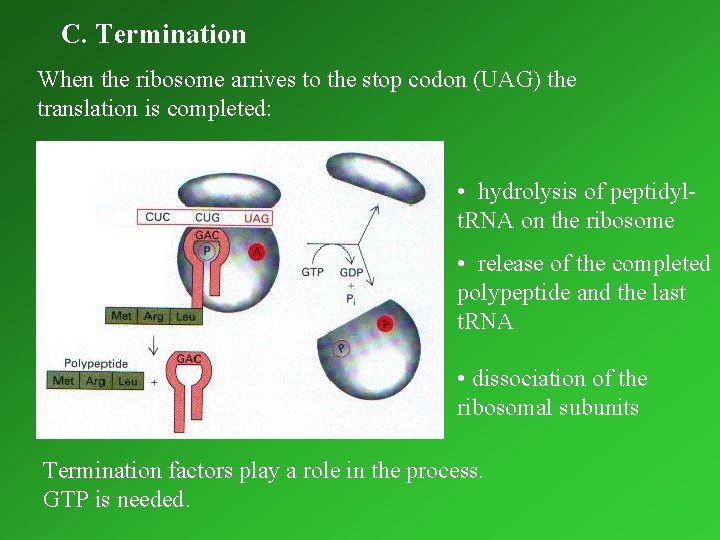
C. Termination When the ribosome arrives to the stop codon (UAG) the translation is completed: • hydrolysis of peptidylt. RNA on the ribosome • release of the completed polypeptide and the last t. RNA • dissociation of the ribosomal subunits Termination factors play a role in the process. GTP is needed.
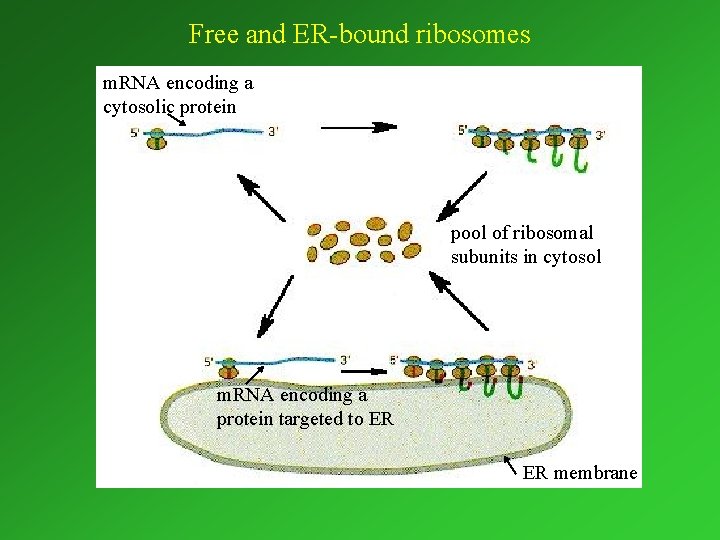
Free and ER-bound ribosomes m. RNA encoding a cytosolic protein pool of ribosomal subunits in cytosol m. RNA encoding a protein targeted to ER ER membrane
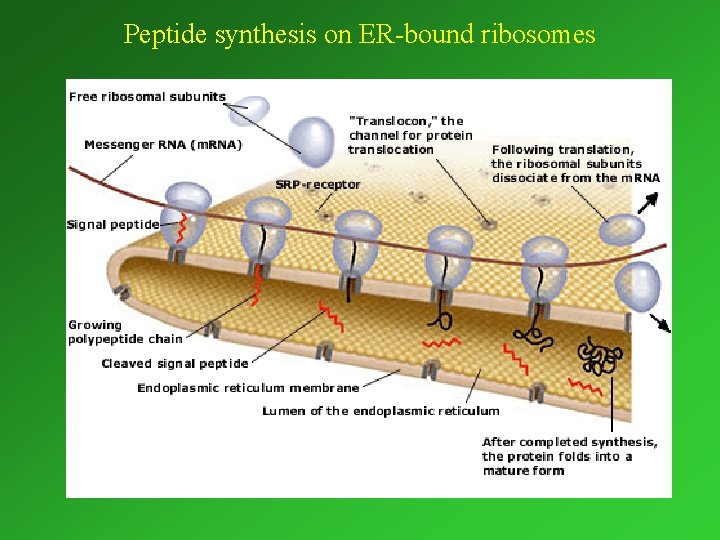
Peptide synthesis on ER-bound ribosomes
Posttranslational modification of proteins After the completiton of translation numerous polypeptides and proteins undergo posttranslational modifications These modifications can influence their structure and function. Most important posttranslational modifications: • specific proteolysis • removal of the first Met • glycosylation • phosphorylation
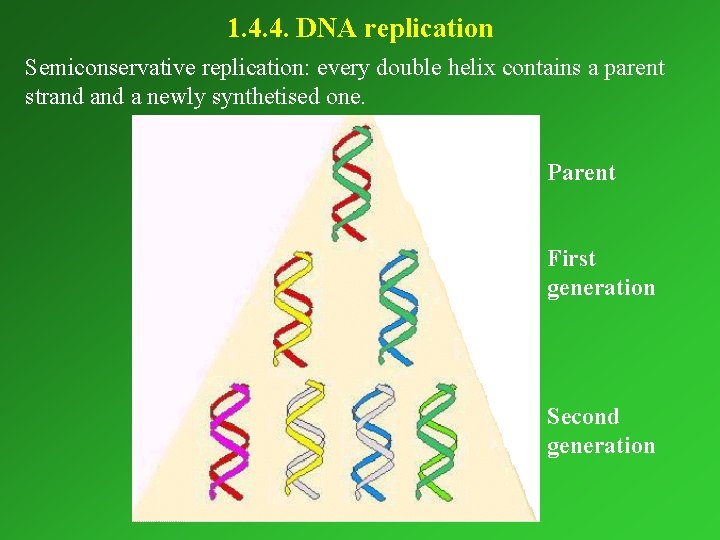
1. 4. 4. DNA replication Semiconservative replication: every double helix contains a parent strand a newly synthetised one. Parent First generation Second generation
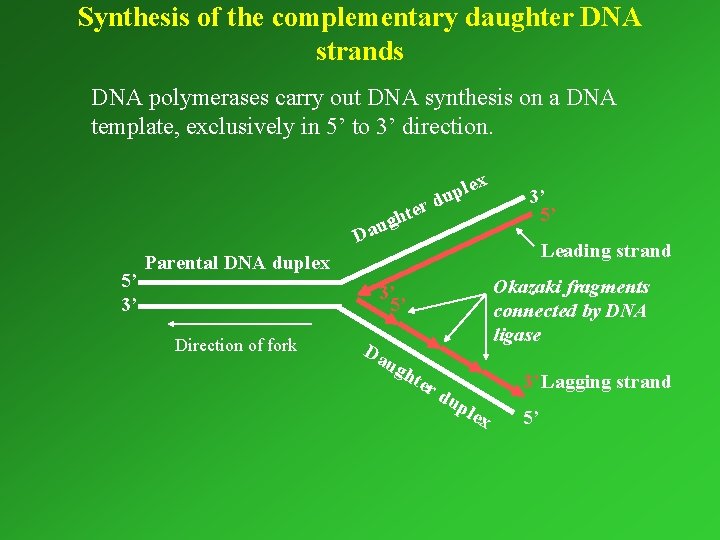
Synthesis of the complementary daughter DNA strands DNA polymerases carry out DNA synthesis on a DNA template, exclusively in 5’ to 3’ direction. ter gh u a D 5’ 3’ lex p u d 3’ 5’ Leading strand Parental DNA duplex Okazaki fragments connected by DNA ligase 3’ 5’ Direction of fork Da u gh ter 3’ Lagging strand du ple x 5’
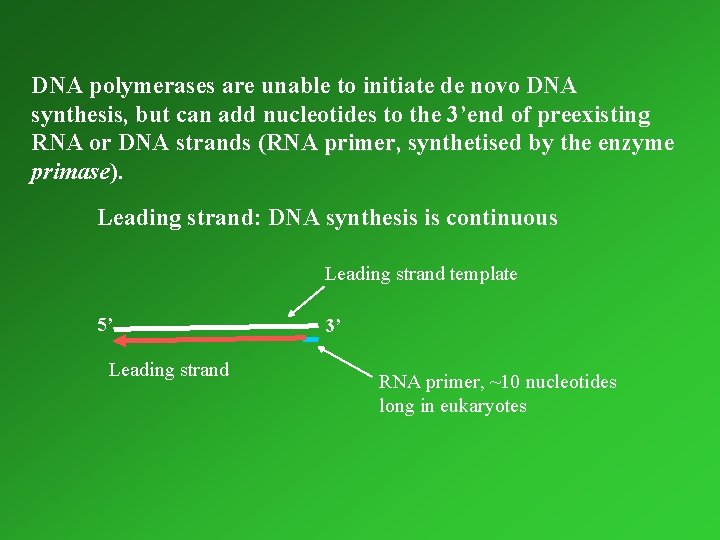
DNA polymerases are unable to initiate de novo DNA synthesis, but can add nucleotides to the 3’end of preexisting RNA or DNA strands (RNA primer, primer synthetised by the enzyme primase). Leading strand: DNA synthesis is continuous Leading strand template 5’ Leading strand 3’ RNA primer, ~10 nucleotides long in eukaryotes
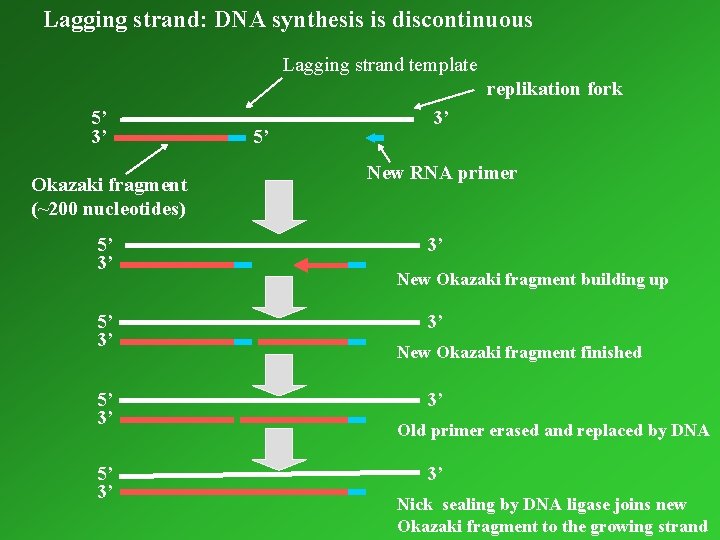
Lagging strand: DNA synthesis is discontinuous Lagging strand template replikation fork 5’ 3’ Okazaki fragment (~200 nucleotides) 5’ 3’ 5’ 3’ New RNA primer 3’ New Okazaki fragment building up 3’ New Okazaki fragment finished 3’ Old primer erased and replaced by DNA 3’ Nick sealing by DNA ligase joins new Okazaki fragment to the growing strand

DNA repair, mutations Maintainig genetic stability requires accurate mechanism of replication as well as repair of lesions that occur continually in DNA. Most spontaneous changes are immediately corrected by the complex process of DNA repair, similarly to replication, relies on basepairing and involves several different pathways. If this process fails, permanent change – mutation – occurs in DNA. Mutations in vital positions of the DNA sequence destroy the organism, others might cause advantageous modifications in the gene products, contributing to the driving force of the evolution.
- Slides: 67