INTRODUCTION TO BIOSTAT LINUX CLUSTER By Helen Wang
INTRODUCTION TO BIOSTAT LINUX CLUSTER By Helen Wang Oct 20, 2017
What is HPC • High-Performance Computing (HPC) clusters are characterized by many cores and processors, lots of memory, high-speed networking, and large data storage – all shared across many rack-mounted servers. User programs that run on a cluster are called jobs, and they are typically managed through a queueing system for optimal utilization of all available resources. • An HPC cluster is made of many separated servers, called nodes, filling into racks. HPC typically involves simulation of numerical models or analysis of data from scientific instrumentation. • Difference between HPC Cluster and Cloud? ? --Cluster is a group of computers connected by a local area network; tightly coupled for function of large computation or single service --Grid and Clouds are more wide scale, loosely coupled, with software and hardware to deliver services to public and private group.
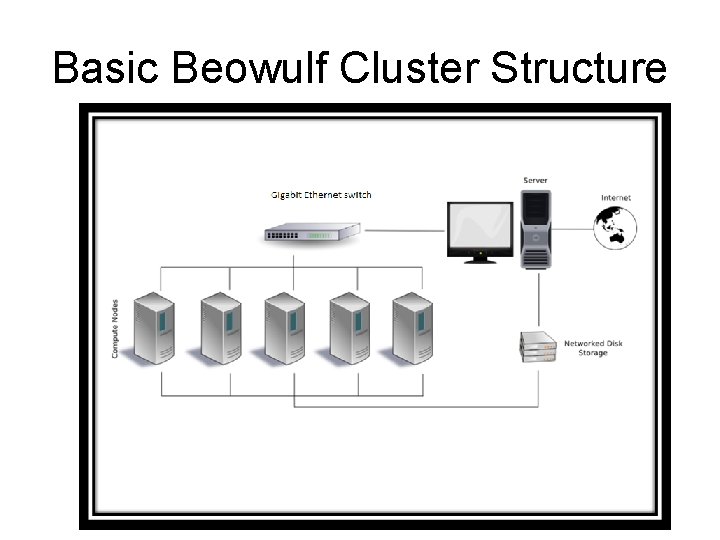
Basic Beowulf Cluster Structure

A brief look of our cluster

What our cluster can do Intensive computation Leveraging a total of 1000+ cores, and large memory available, the Linux cluster system is configured to ensure multiple job types run efficiently. Two types of computations: Serial and Parallel Serial Computing: Traditionally, software has been written for serial computation: • A problem is broken into a discrete series of instructions • Instructions are executed sequentially one after another • Executed on a single processor • Only one instruction may execute at any moment in time

Serial computing • For example
Parallel Computing • • • In the simplest sense, parallel computing is the simultaneous use of multiple compute resources to solve a computational problem: A problem is broken into discrete parts that can be solved concurrently Each part is further broken down to a series of instructions Instructions from each part execute simultaneously on different processors An overall control/coordination mechanism is employed The computational problem should be able to: • • • Be broken apart into discrete pieces of work that can be solved simultaneously; Execute multiple program instructions at any moment in time; Be solved in less time with multiple compute resources than with a single compute resource. • The compute resources are typically: • A single computer with multiple processors/cores • An arbitrary number of such computers connected by a network Embarrassingly Parallel • Solving many similar, but independent tasks simultaneously; little to no need for coordination between the tasks.
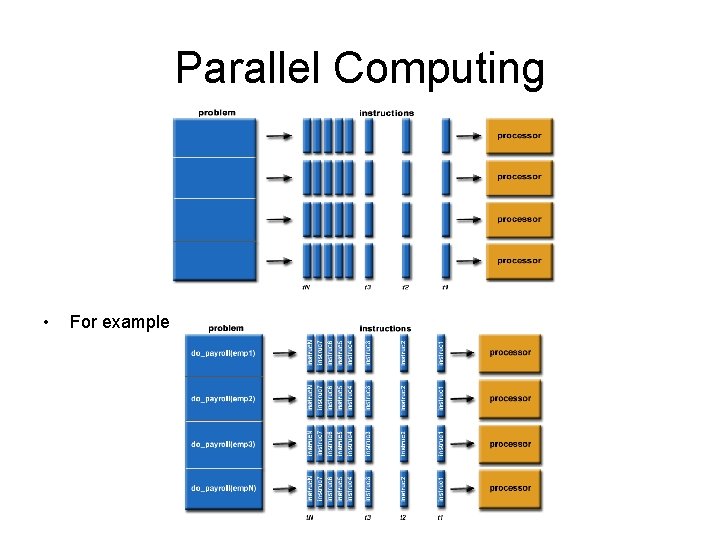
Parallel Computing • For example

why we use cluster and what to expect WHY • Use it when you have lots of independent programs to run for a project – think about parallel • Use it when your program is taking very long time to finish –give your laptop a break ( normally more than 3 -4 hours, you should consider using cluster) • Use it when your program is using up lot of memory – to avoid crash on your pc What to expect • It is not going to be super fast to run a single command line even with hundreds of cpu cores • It is not set up for one single user even it looks like one super computer to you • You are sharing resources with lots of hardcore users
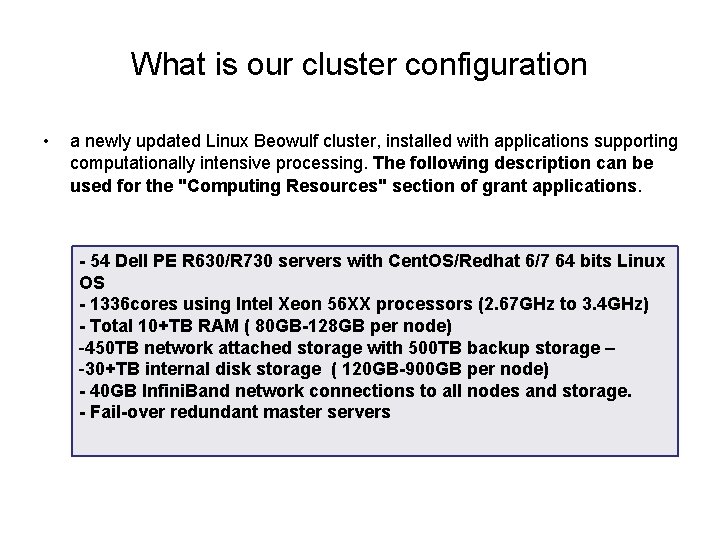
What is our cluster configuration • a newly updated Linux Beowulf cluster, installed with applications supporting computationally intensive processing. The following description can be used for the "Computing Resources" section of grant applications. - 54 Dell PE R 630/R 730 servers with Cent. OS/Redhat 6/7 64 bits Linux OS - 1336 cores using Intel Xeon 56 XX processors (2. 67 GHz to 3. 4 GHz) - Total 10+TB RAM ( 80 GB-128 GB per node) -450 TB network attached storage with 500 TB backup storage – -30+TB internal disk storage ( 120 GB-900 GB per node) - 40 GB Infini. Band network connections to all nodes and storage. - Fail-over redundant master servers

Software available on cluster • • • R 3. 4. 1 with CRAN packages and BIOCONDUCTOR packages C++/G++ and Fortran compilers JAVA compiler Perl compiler Python 3. 5 and Biopython compiler SAS-9. 4 for Linux 64 bit PLINK MPI and Open. MPI PBS Pro 12. 2 (a portable batch system for cluster) Additional open source software upon user request

Who can use it and when? • The cluster supports department faculty, research affiliates and students working on intensive computation in both serial and parallel processing. All users are able to access cluster through VCU VPN for a secured connection. • You can use it at anytime and anywhere, mostly when you start your graduate dissertation. Please consider start your computation work on cluster for at least 6 months in advanced, the system can be very busy and you won’t get the sufficient resources for estimated time. • Come to see me if you need help preparing the code for parallel processing.
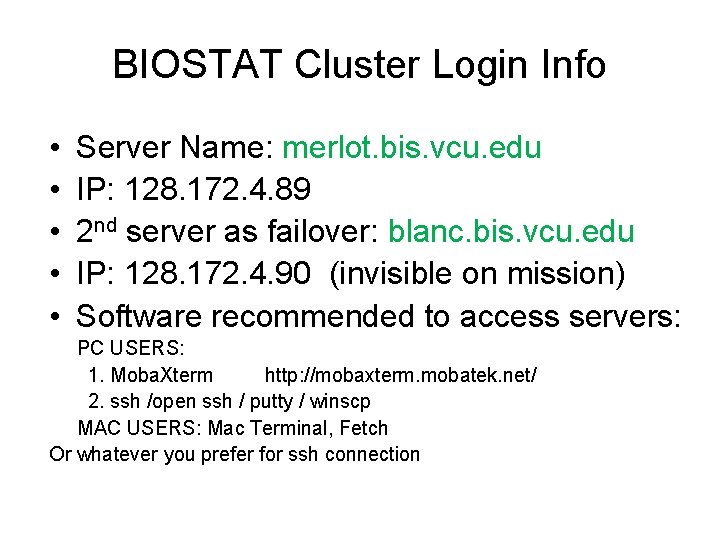
BIOSTAT Cluster Login Info • • • Server Name: merlot. bis. vcu. edu IP: 128. 172. 4. 89 2 nd server as failover: blanc. bis. vcu. edu IP: 128. 172. 4. 90 (invisible on mission) Software recommended to access servers: PC USERS: 1. Moba. Xterm http: //mobaxterm. mobatek. net/ 2. ssh /open ssh / putty / winscp MAC USERS: Mac Terminal, Fetch Or whatever you prefer for ssh connection

Access Cluster Server and nodes Master node (master 1 / master 2): merlot. bis. vcu. edu Running Cent. OS ( redhat kenrnel)Version 6. 8, x 86 -64 Open source or Software download – choose 64 bits Cent. OS or RHEL 6 or scientific Linux if possible Purposes: front-end user interface; slow; - not for running any jobs. Jobs running on master will be terminated without notice accessible from outside through VPN; Slave nodes (nodes): node 1–node 54 : dual multiple cores Intel Xeon processors with 128 -512 GB GB RAM Purposes: computation; not prefer to access user interface, accessible via master and managed by portable batch management ( PBS ); fast; internal network; -10. 0. 10. X, not accessible directly from outside
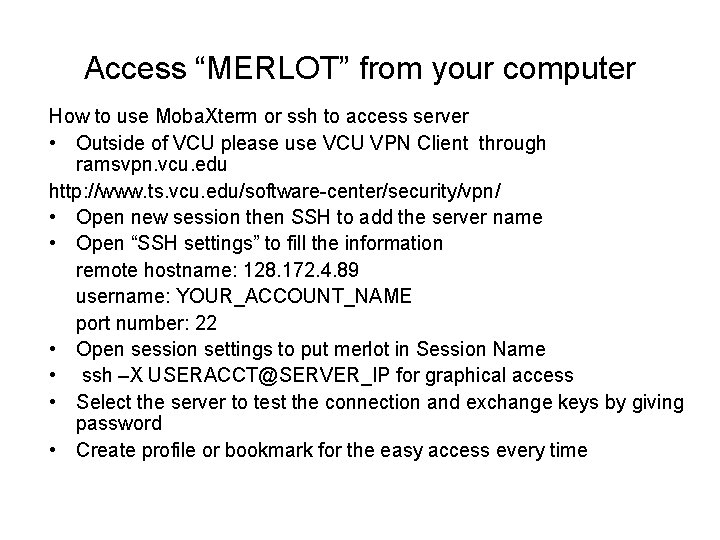
Access “MERLOT” from your computer How to use Moba. Xterm or ssh to access server • Outside of VCU please use VCU VPN Client through ramsvpn. vcu. edu http: //www. ts. vcu. edu/software-center/security/vpn/ • Open new session then SSH to add the server name • Open “SSH settings” to fill the information remote hostname: 128. 172. 4. 89 username: YOUR_ACCOUNT_NAME port number: 22 • Open session settings to put merlot in Session Name • ssh –X USERACCT@SERVER_IP for graphical access • Select the server to test the connection and exchange keys by giving password • Create profile or bookmark for the easy access every time

UNIX Commands You Need to Know • • • • • • • pwd clear mkdir cp cd mv ls head more less wc man rm chmod grep with head and tail nano sed cut top scp acct 1@server 1: /yourpath/yourfiles acct 2@server 2: /yourpath/ nproc –all lscpu cat /proc/cpuinfo echo Cores = $(( $(lscpu | awk '/Socket/{ print $2 }') * $(lscpu | awk '/Core/{ print $4 }') )) vmstat -s | grep memory cat /proc/meminfo | grep Mem. Total | awk '{ print $2 }’ awk '/Mem. Total/ {print $2}' /proc/meminfo

How to use cluster to submit jobs IMPORTANT • The logon machine (MERLOT. BIS. VCU. EDU) is only used for login. Jobs running on light will be terminated without notice. What do you need to submit a job via PBS • an executable script, the script can be a program code ( R, SAS or other language) , shell script or a collection of command lines. • test how much resources and time you may need before you submit multiple jobs. • which queue to use to submit a job Node and queue configuration • Nodes: Nodes are the physical computer servers incorporated together to make the cluster. • Queues: Queues are being used by pbs scheduler to send jobs to different nodes, each node is assigned to a queue to handle different type of jobs. • Most queue limits can be checked by running the command qstat -q. Note that if you need more job permissions, please send request to system admin and your supervisor to get a temporary expansion on job submissions
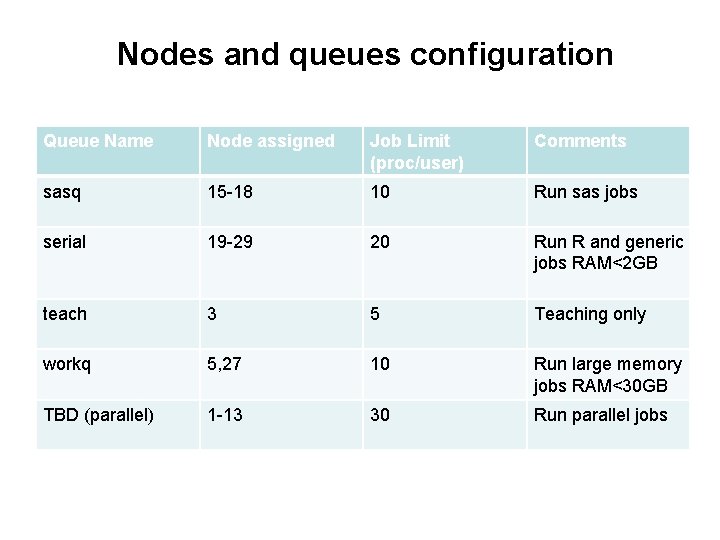
Nodes and queues configuration Queue Name Node assigned Job Limit (proc/user) Comments sasq 15 -18 10 Run sas jobs serial 19 -29 20 Run R and generic jobs RAM<2 GB teach 3 5 Teaching only workq 5, 27 10 Run large memory jobs RAM<30 GB TBD (parallel) 1 -13 30 Run parallel jobs

Submitting a Jobs are submitted to a PBS queue so that PBS can dispatch them to be run on one or more of a cluster's compute nodes. There are two main types of PBS jobs: • Non-interactive Batch Jobs: This is the most common PBS job. A job script is created that contains PBS resource requests and the commands necessary to execute the job. The job script is then submitted to PBS to be run non-interactively. • Interactive Batch Jobs: This is a way to get an interactive terminal on one or more of the compute nodes of a cluster. Commands can then be run interactively through that terminal directly on the compute nodes for the duration of the job. Interactive jobs are helpful for such things as program debugging and running many short jobs.

A PBS script is a standard Unix/Linux shell script that contains a few extra comments at the beginning that specify directives to PBS. These comments all begin with #PBS. • The most important PBS directives are: PBS Directives Description #PBS –I walltime=HH: MM: SS This directive specifies the maximum walltime (real time, not CPU time) that a job should take. If this limit is exceeded, PBS will stop the job. Keeping this limit close to the actual expected time of a job can allow a job to start more quickly than if the maximum walltime is always requested. #PBS -l pmem=SIZEgb This directive specifies the maximum amount of physical memory used by any process in the job. For example, if the job would run four processes and each would use up to 2 GB (gigabytes) of memory, then the directive would read #PBS -l pmem=2 gb #PBS -l nodes=N: ppn=M This specifies the number of nodes (nodes=N) and the number of processors per node (ppn=M) that the job should use. PBS treats a processor core as a processor, so a system with eight cores per compute node can have ppn=8 as its maximum ppn request. Note that unless a job has some inherent parallelism of its own through something like MPI or Open. MPI, requesting more than a single processor on a single node is usually wasteful and can impact the job start time. #PBS -q queuename This specifies what PBS queue a job should be submitted to. This is only necessary if a user has access to a special queue. This option can and should be omitted for jobs being submitted to a system's default queue. #PBS -j oe Normally when a command runs it prints its output to the screen. This output is often normal output and error output. This directive tells PBS to put both normal output and error output into the same output file.

An example of PBS script #This is a sample PBS script. It will request 1 processor on 1 node for 10 hours. # #Request 1 processors on 1 node # #PBS -l nodes=1: ppn=1 # #Request 10 hours of walltime # #PBS -l walltime=10: 00 # #Request 1 gigabyte of memory per process # #PBS -l mem=1 gb # #Request that regular output and terminal output go to the same file # #PBS -j oe # #The following is the body of the script. By default, PBS scripts execute in your home directory, not the #directory from which they were submitted. The following line places you in the directory from which the job #was submitted. # cd $PBS_O_WORKDIR # #Now we want to run the program "hello" is in the directory that this script is being submitted from, #$PBS_O_WORKDIR. # echo " " echo "Job started on `hostname` at `date`". /hello echo " " echo "Job Ended at `date`" echo " "

Template used on cluster • Modify template to create your own pbs script for running programs #!/bin/bash #PBS -q serial #PBS -N MYSCRIPT # # # cd to the directory from which I submitted the job. Otherwise it will execute in my home directory. # set WORKDIR = ~/YOURWORDIR #PBS -V #echo “PBS batch job id is $PBS_JOBID“ echo "Working directory of this job is: " $WORKDIR # echo "Beginning to run job“ Command line you need to execute the job ( /home/huan/bin/calculate PARAMETEERS)

• Job Submission Syntax qsub SCRIPTFILE Existing job submission scripts – – – /usr/local/bin/q* R USERS q. R YOUR_R_SCRIPT Large memory R jobs q. RL YOUR_R_SCRIPT SAS USERS qsas YOUR_SAS_CODE Generic or other resources qsub YOUR_OWN_SCRIPT Interactive Batch Jobs • Interactive PBS jobs are similar to non-interactive PBS jobs in that they are submitted to PBS via the command qsub. When submitting an interactive PBS job, PBS script is not necessary. All PBS directives can be specified on the command line. The syntax for qsub for submitting an interactive PBS job is: qsub -I. . . pbs directives. . • The -I flag above tells qsub that this is an interactive job. • The following example shows using qsub to submit an interactive job using one processor on one node for four hours merlot: ~$ qsub -I -l nodes=1: ppn=1 -l walltime=4: 00 qsub: waiting for job 1064159. merlot. bis. vcu. edu start qsub: job 1064159. merlot. bis. vcu. edu ready node 12: ~$ • There are two things of note here. The first is that the qsub command doesn't exit when run with the interactive -I flag. Instead, it waits until the job is started and gives a prompt on the first compute node assigned to a job. The second thing of note is the prompt node 12: ~$ - this shows that commands are now being executed on the compute node 12.
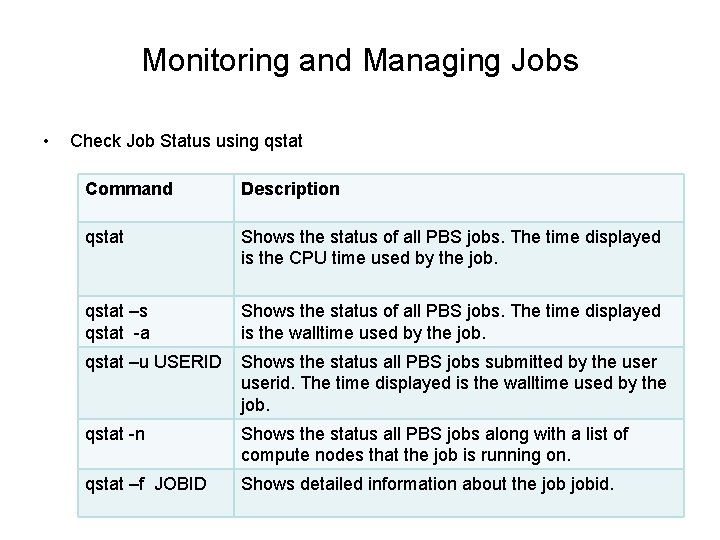
Monitoring and Managing Jobs • Check Job Status using qstat Command Description qstat Shows the status of all PBS jobs. The time displayed is the CPU time used by the job. qstat –s qstat -a Shows the status of all PBS jobs. The time displayed is the walltime used by the job. qstat –u USERID Shows the status all PBS jobs submitted by the userid. The time displayed is the walltime used by the job. qstat -n Shows the status all PBS jobs along with a list of compute nodes that the job is running on. qstat –f JOBID Shows detailed information about the jobid.

Job Running Status State meaning Q The job is queued and is waiting to start. R The job is currently running E The job is currently ending. H The job has a user or system hold on it and will not be eligible to run until the hold is removed.

Managing jobs • Deleting jobs - qdel JOBID delete a job by Job_ID - qdel $(qselect –u USERNAME) delete all jobs owned by USERNAME • View job output If the PBS directive #PBS -j oe is used in a PBS script, the non-error and the error output are both written to the Jobname. o. Job_ID file. Job. Name. o. Job. ID : This file would contain the non-error output that would normally be written to the screen. Job. Name. e. Job. ID: This file would contain the error output that would normally be written to the screen.
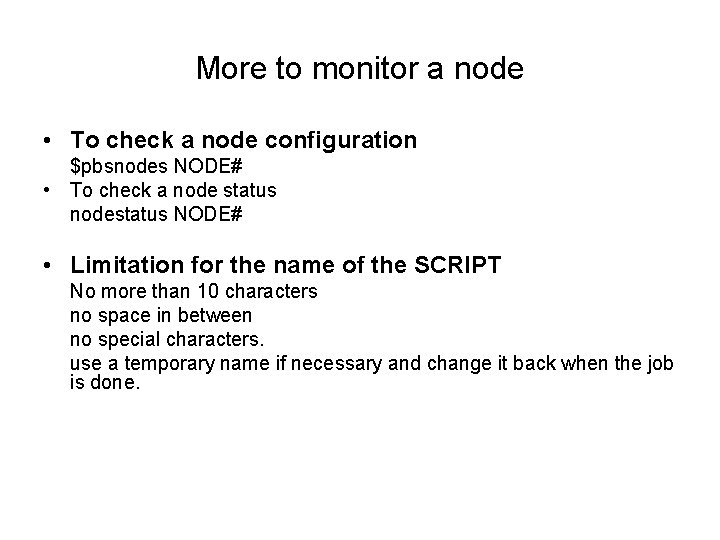
More to monitor a node • To check a node configuration $pbsnodes NODE# • To check a node status nodestatus NODE# • Limitation for the name of the SCRIPT No more than 10 characters no space in between no special characters. use a temporary name if necessary and change it back when the job is done.

Use existing scripts to submit jobs • Scripts for R jobs submission /usr/local/bin/q. R, q. Rh, qsas R public libraries /home/R/Rlib-3. 4. 1 Python public library /home/python-site-packages

At Last • Edit file using nano or vi http: //www. ts. vcu. edu/askit/research/unix-for-researchers-atvcu/unix-text-editors/the-pico-editor/ http: //www. ts. vcu. edu/askit/research/unix-for-researchers-atvcu/unix-text-editors/the-vi-text-editor/ • use samba connection to map a network drive on PC, recommending to use “Edit. Pad Lite” • Useful links http: //www. ts. vcu. edu/askit/research/unix-for-researchers-atvcu/unix-survival-guide-a-user-manual/ • Wiki page for biostat cluster – need vcu e. ID to login • https: //wiki. vcu. edu/display/biosit/Cluster+Computing
- Slides: 29