Introduction to bioinformatics Lecture 9 Multiple sequence alignment
Introduction to bioinformatics Lecture 9 Multiple sequence alignment (3)
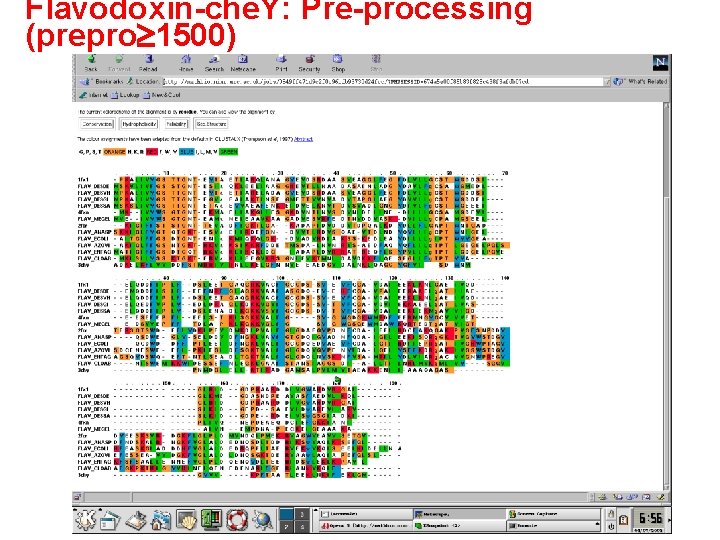
Flavodoxin-che. Y: Pre-processing (prepro 1500)
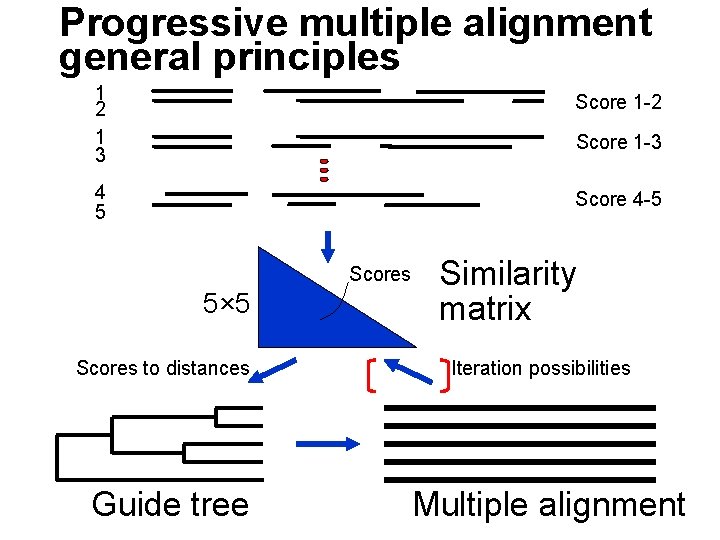
Progressive multiple alignment general principles 1 2 1 3 Score 1 -2 4 5 Score 4 -5 Score 1 -3 Scores 5× 5 Scores to distances Guide tree Similarity matrix Iteration possibilities Multiple alignment
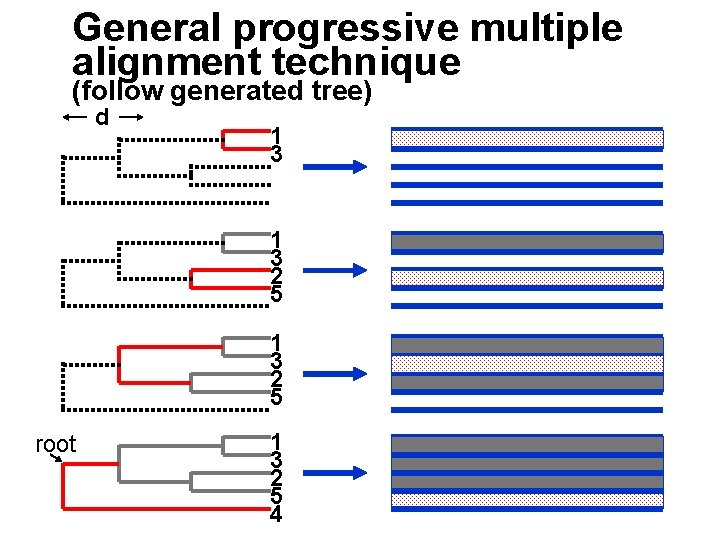
General progressive multiple alignment technique (follow generated tree) d 1 3 2 5 root 1 3 2 5 4
Strategies for multiple sequence alignment n n Profile pre-processing Secondary structure-induced alignment Globalised local alignment Matrix extension Objective: integrate secondary structure information to anchor alignments and avoid errors
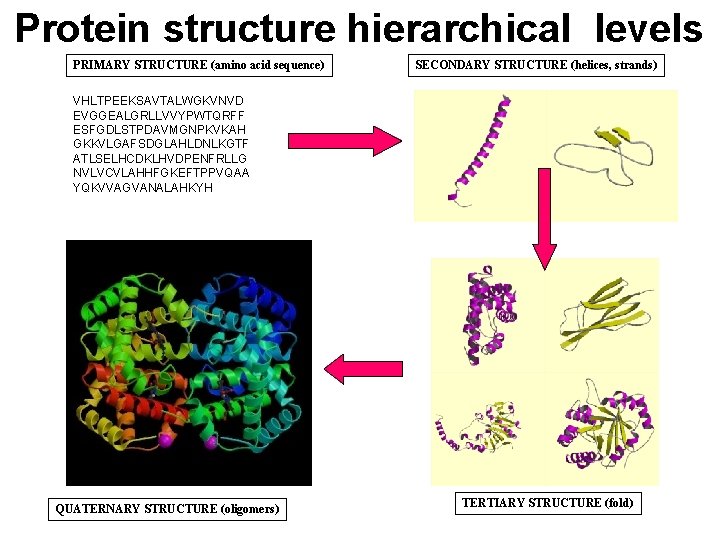
Protein structure hierarchical levels PRIMARY STRUCTURE (amino acid sequence) SECONDARY STRUCTURE (helices, strands) VHLTPEEKSAVTALWGKVNVD EVGGEALGRLLVVYPWTQRFF ESFGDLSTPDAVMGNPKVKAH GKKVLGAFSDGLAHLDNLKGTF ATLSELHCDKLHVDPENFRLLG NVLVCVLAHHFGKEFTPPVQAA YQKVVAGVANALAHKYH QUATERNARY STRUCTURE (oligomers) TERTIARY STRUCTURE (fold)

Why use (predicted) structural information n “Structure more conserved than sequence” u n n Many structural protein families (e. g. globins) have family members with very low sequence similarities. For example, globin sequences identities can be as low as 10% while still having an identical fold. This means that you can still observe equivalent secondary structures in homologous proteins even if sequence similarities are extremely low. But you are dependent on the quality of prediction methods. For example, secondary structure prediction is currently at 76% correctness. So, 1 out of 4 predicted amino acids is still incorrect.
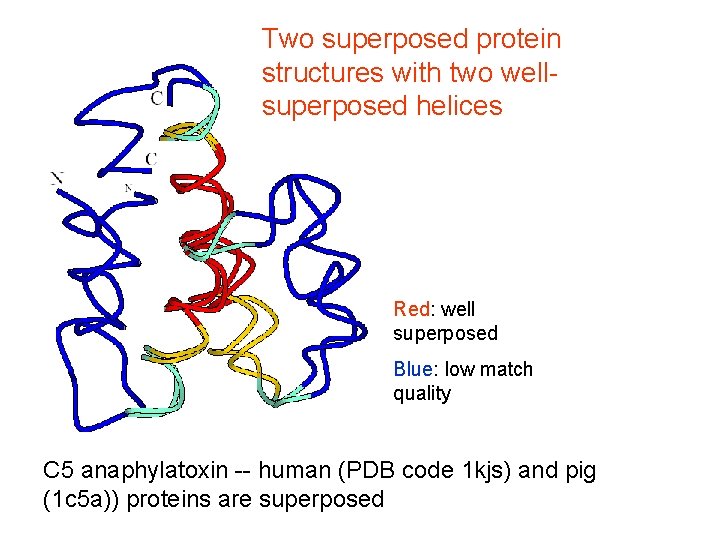
Two superposed protein structures with two wellsuperposed helices Red: well superposed Blue: low match quality C 5 anaphylatoxin -- human (PDB code 1 kjs) and pig (1 c 5 a)) proteins are superposed
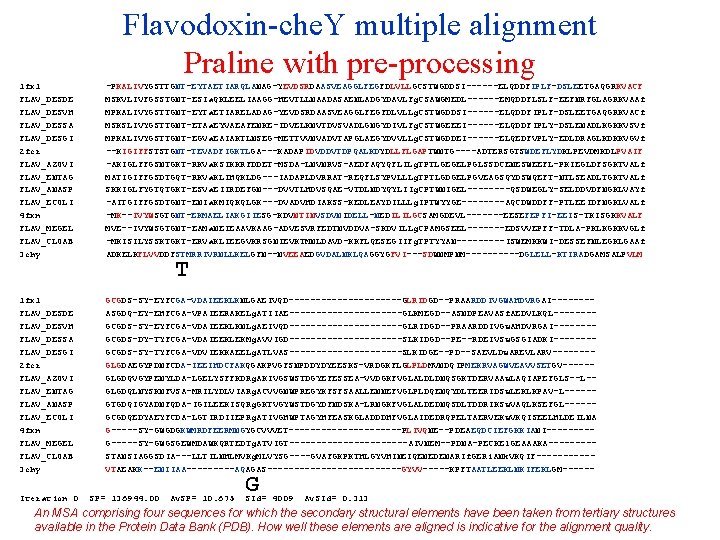
Flavodoxin-che. Y multiple alignment Praline with pre-processing 1 fx 1 FLAV_DESDE FLAV_DESVH FLAV_DESSA FLAV_DESGI 2 fcr FLAV_AZOVI FLAV_ENTAG FLAV_ANASP FLAV_ECOLI 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy -PKALIVYGSTTGNT-EYTAETIARQLANAG-YEVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSI------ELQDDFIPLF-DSLEETGAQGRKVACF MSKVLIVFGSSTGNT-ESIa. QKLEELIAAGG-HEVTLLNAADASAENLADGYDAVLFg. CSAWGMEDL------EMQDDFLSLF-EEFNRFGLAGRKVAAf MPKALIVYGSTTGNT-EYTa. ETIARELADAG-YEVDSRDAASVEAGGLFEGFDLVLLg. CSTWGDDSI------ELQDDFIPLF-DSLEETGAQGRKVACf MSKSLIVYGSTTGNT-ETAa. EYVAEAFENKE-IDVELKNVTDVSVADLGNGYDIVLFg. CSTWGEEEI------ELQDDFIPLY-DSLENADLKGKKVSVf MPKALIVYGSTTGNT-EGVa. EAIAKTLNSEG-METTVVNVADVTAPGLAEGYDVVLLg. CSTWGDDEI------ELQEDFVPLY-EDLDRAGLKDKKVGVf --KIGIFFSTSTGNT-TEVADFIGKTLGA---KADAPIDVDDVTDPQALKDYDLLFLGAPTWNTG----ADTERSGTSWDEFLYDKLPEVDMKDLPVAIF -AKIGLFFGSNTGKT-RKVa. KSIKKRFDDET-MSDA-LNVNRVS-AEDFAQYQFLILg. TPTLGEGELPGLSSDCENESWEEFL-PKIEGLDFSGKTVALf MATIGIFFGSDTGQT-RKVa. KLIHQKLDG---IADAPLDVRRAT-REQFLSYPVLLLg. TPTLGDGELPGVEAGSQYDSWQEFT-NTLSEADLTGKTVALf SKKIGLFYGTQTGKT-ESVa. EIIRDEFGN---DVVTLHDVSQAE-VTDLNDYQYLIIg. CPTWNIGEL----QSDWEGLY-SELDDVDFNGKLVAYf -AITGIFFGSDTGNT-ENIa. KMIQKQLGK---DVADVHDIAKSS-KEDLEAYDILLLg. IPTWYYGE----AQCDWDDFF-PTLEEIDFNGKLVALf -MK--IVYWSGTGNT-EKMAELIAKGIIESG-KDVNTINVSDVNIDELL-NEDILILGCSAMGDEVL-------EESEFEPFI-EEIS-TKISGKKVALF MVE--IVYWSGTGNT-EAMa. NEIEAAVKAAG-ADVESVRFEDTNVDDVA-SKDVILLg. CPAMGSEEL-------EDSVVEPFF-TDLA-PKLKGKKVGLf -MKISILYSSKTGKT-ERVa. KLIEEGVKRSGNIEVKTMNLDAVD-KKFLQESEGIIFg. TPTYYAN-----ISWEMKKWI-DESSEFNLEGKLGAAf ADKELKFLVVDDFSTMRRIVRNLLKELGFN--NVEEAEDGVDALNKLQAGGYGFVI---SDWNMPNM-----DGLELL-KTIRADGAMSALPVLM 1 fx 1 FLAV_DESDE FLAV_DESVH FLAV_DESSA FLAV_DESGI 2 fcr FLAV_AZOVI FLAV_ENTAG FLAV_ANASP FLAV_ECOLI 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy GCGDS-SY-EYFCGA-VDAIEEKLKNLGAEIVQD-----------GLRIDGD--PRAARDDIVGWAHDVRGAI-------ASGDQ-EY-EHFCGA-VPAIEERAKELg. ATIIAE-----------GLKMEGD--ASNDPEAVASf. AEDVLKQL-------GCGDS-SY-EYFCGA-VDAIEEKLKNLg. AEIVQD-----------GLRIDGD--PRAARDDIVGw. AHDVRGAI-------GCGDS-DY-TYFCGA-VDAIEEKLEKMg. AVVIGD-----------SLKIDGD--PE--RDEIVSw. GSGIADKI-------GCGDS-SY-TYFCGA-VDVIEKKAEELg. ATLVAS-----------SLKIDGE--PD--SAEVLDw. AREVLARV-------GLGDAEGYPDNFCDA-IEEIHDCFAKQGAKPVGFSNPDDYDYEESKS-VRDGKFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-----GLGDQVGYPENYLDA-LGELYSFFKDRg. AKIVGSWSTDGYEFESSEA-VVDGKFVGLALDLDNQSGKTDERVAAw. LAQIAPEFGLS--L-GLGDQLNYSKNFVSA-MRILYDLVIARg. ACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSw. LEKLKPAV-L-----GTGDQIGYADNFQDA-IGILEEKISQRg. GKTVGYWSTDGYDFNDSKA-LRNGKFVGLALDEDNQSDLTDDRIKSw. VAQLKSEFGL-----GCGDQEDYAEYFCDA-LGTIRDIIEPRg. ATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKw. VKQISEELHLDEILNA G-----SY-GWGDGKWMRDFEERMNGYGCVVVET-----------PLIVQNE--PDEAEQDCIEFGKKIANI----G-----SY-GWGSGEWMDAWKQRTEDTg. ATVIGT-----------AIVNEM--PDNA-PECKEl. GEAAAKA----STANSIAGGSDIA---LLTILNHLMVKg. MLVYSG----GVAFGKPKTHLGYVHINEIQENEDENARIf. GERi. ANk. VKQIF-----VTAEAKK--ENIIAA-----AQAGAS-------------GYVV-----KPFTAATLEEKLNKIFEKLGM------ Iteration 0 T SP= 136944. 00 Av. SP= 10. 675 G SId= 4009 Av. SId= 0. 313 An MSA comprising four sequences for which the secondary structural elements have been taken from tertiary structures available in the Protein Data Bank (PDB). How well these elements are aligned is indicative for the alignment quality.

Secondary structureinduced alignment iteration
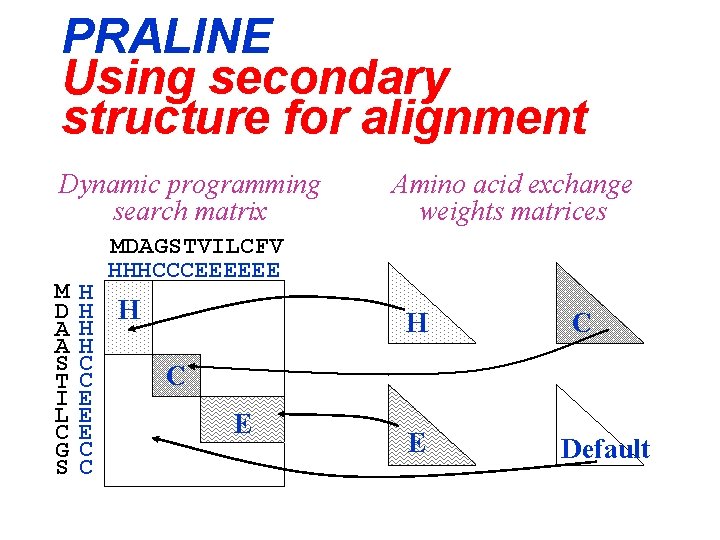
PRALINE Using secondary structure for alignment Dynamic programming search matrix M D A A S T I L C G S Amino acid exchange weights matrices MDAGSTVILCFV HHHCCCEEEEEE H H H C C E E E C C H C C E E Default
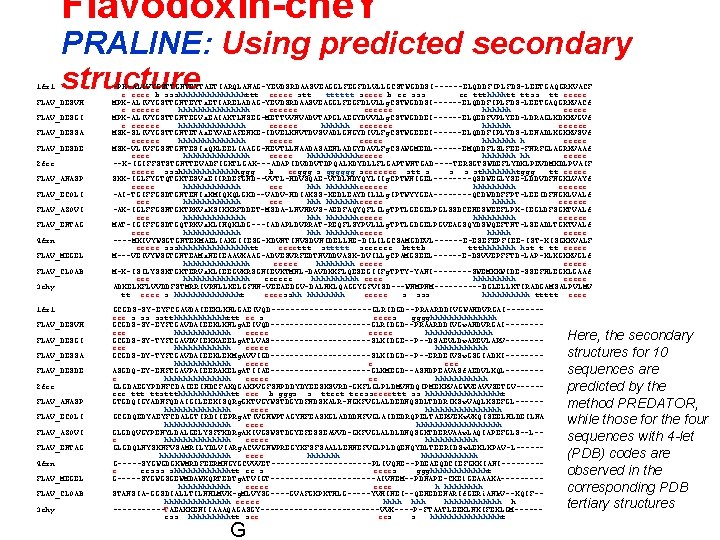
Flavodoxin-che. Y 1 fx 1 PRALINE: Using predicted secondary structure FLAV_DESVH FLAV_DESGI FLAV_DESSA FLAV_DESDE 2 fcr FLAV_ANASP FLAV_ECOLI FLAV_AZOVI FLAV_ENTAG 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy 1 fx 1 FLAV_DESVH FLAV_DESGI FLAV_DESSA FLAV_DESDE 2 fcr FLAV_ANASP FLAV_ECOLI FLAV_AZOVI FLAV_ENTAG 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy -PK-ALIVYGSTTGNTEYTAETIARQLANAG-YEVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSI------ELQDDFIPLFDS-LEETGAQGRKVACF e eeee b ssshhhhhhhttt eeeee stt tttttt seeee b ee sss ee ttthhhhtt ttss tt eeeee MPK-ALIVYGSTTGNTEYTa. ETIARELADAG-YEVDSRDAASVEAGGLFEGFDLVLLg. CSTWGDDSI------ELQDDFIPLFDS-LEETGAQGRKVACf e eeeeee hhhhhhhh eeeeee hhhhhh eeeee MPK-ALIVYGSTTGNTEGVa. EAIAKTLNSEG-METTVVNVADVTAPGLAEGYDVVLLg. CSTWGDDEI------ELQEDFVPLYED-LDRAGLKDKKVGVf e eeeeee hhhhhhh eeeeee MSK-SLIVYGSTTGNTETAa. EYVAEAFENKE-IDVELKNVTDVSVADLGNGYDIVLFg. CSTWGEEEI------ELQDDFIPLYDS-LENADLKGKKVSVf eeeeee hhhhhhh h eeeee MSK-VLIVFGSSTGNTESIa. QKLEELIAAGG-HEVTLLNAADASAENLADGYDAVLFg. CSAWGMEDL------EMQDDFLSLFEE-FNRFGLAGRKVAAf eeee hhhhhhh eeeee hhhhhheeeee hhhhhhh hh eeeee --K-IGIFFSTSTGNTTEVADFIGKTLGAK---ADAPIDVDDVTDPQALKDYDLLFLGAPTWNTGAD----TERSGTSWDEFLYDKLPEVDMKDLPVAIF eeeee ssshhhhhhhggg b eeggg s gggggg seeeeeee stt s sthhhhhhhtggg tt eeeee SKK-IGLFYGTQTGKTESVa. EIIRDEFGND--VVTL-HDVSQAE-VTDLNDYQYLIIg. CPTWNIGEL----QSDWEGLYSE-LDDVDFNGKLVAYf eeeee hhhhhhheeeeee hhhhh eeeeee -AI-TGIFFGSDTGNTENIa. KMIQKQLGKD--VADV-HDIAKSS-KEDLEAYDILLLg. IPTWYYGEA----QCDWDDFFPT-LEEIDFNGKLVALf eee hhhhhhheeeee hhhhh eeeeee -AK-IGLFFGSNTGKTRKVa. KSIKKRFDDET-MSDA-LNVNRVS-AEDFAQYQFLILg. TPTLGEGELPGLSSDCENESWEEFLPK-IEGLDFSGKTVALf eee hhhhhhheeeee hhhhh eeeeee MAT-IGIFFGSDTGQTRKVa. KLIHQKLDG---IADAPLDVRRAT-REQFLSYPVLLLg. TPTLGDGELPGVEAGSQYDSWQEFTNT-LSEADLTGKTVALf eeee hhhhhhheeeee hhhhh eeeee ----MKIVYWSGTGNTEKMAELIAKGIIESG-KDVNTINVSDVNIDELLNE-DILILGCSAMGDEVL------E-ESEFEPFIEE-IST-KISGKKVALF eeeee ssshhhhhhhhtt eeeettt sttttt seeeeee btttb ttthhhhhhh hst t tt eeeee M---VEIVYWSGTGNTEAMa. NEIEAAVKAAG-ADVESVRFEDTNVDDVASK-DVILLg. CPAMGSEEL------E-DSVVEPFFTD-LAP-KLKGKKVGLf hhhhhhh eeeee M-K-ISILYSSKTGKTERVa. KLIEEGVKRSGNIEVKTMNL-DAVDKKFLQESEGIIFg. TPTY-YANI----SWEMKKWIDE-SSEFNLEGKLGAAf eee hhhhhhh eeeeee hhhhh eeeee ADKELKFLVVDDFSTMRRIVRNLLKELGFNN-VEEAEDGV-DALNKLQAGGYGFVISD---WNMPNM-----DGLELLKTIRADGAMSALPVLMV tt eeee s hhhhhhht eeeesshh hhhh eeeee s sss hhhhh ttttt eeee GCGDS-SY-EYFCGAVDAIEEKLKNLGAEIVQD-----------GLRIDGD--PRAARDDIVGWAHDVRGAI-------eee s ss sstthhhhhhttt ee s eeees gggghhhhhhh GCGDS-SY-EYFCGAVDAIEEKLKNLg. AEIVQD-----------GLRIDGD--PRAARDDIVGw. AHDVRGAI-------eee hhhhhh eeeee hhhhhhh GCGDS-SY-TYFCGAVDVIEKKAEELg. ATLVAS-----------SLKIDGE--P--DSAEVLDw. AREVLARV-------eee hhhhhh eeeee hhhhhh GCGDS-DY-TYFCGAVDAIEEKLEKMg. AVVIGD-----------SLKIDGD--P--ERDEIVSw. GSGIADKI-------hhhhhh eeeee e eee ASGDQ-EY-EHFCGAVPAIEERAKELg. ATIIAE-----------GLKMEGD--ASNDPEAVASf. AEDVLKQL-------e hhhhhhh eeeee ee hhhhhh GLGDAEGYPDNFCDAIEEIHDCFAKQGAKPVGFSNPDDYDYEESKSVRD-GKFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-----eee ttt ttsttthhhhhhtt eee b gggs s tteet teesseeeettt ss hhhhhhhht GTGDQIGYADNFQDAIGILEEKISQRg. GKTVGYWSTDGYDFNDSKALR-NGKFVGLALDEDNQSDLTDDRIKSw. VAQLKSEFGL-----hhhhhhh eeee hhhhhhhh GCGDQEDYAEYFCDALGTIRDIIEPRg. ATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKw. VKQISEELHLDEILNA hhhhhhh eeee hhhhhhhhh GLGDQVGYPENYLDALGELYSFFKDRg. AKIVGSWSTDGYEFESSEAVVD-GKFVGLALDLDNQSGKTDERVAAw. LAQIAPEFGLS--L-e hhhhhhh eeeee hhhhhh GLGDQLNYSKNFVSAMRILYDLVIARg. ACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSw. LEKLKPAV-L-----hhhhhhhh eeee hhhhhhhhhhhh G-----SYGWGDGKWMRDFEERMNGYGCVVVET-----------PLIVQNE--PDEAEQDCIEFGKKIANI----e eesss shhhhhhtt ee s eeees ggghhhhhht G-----SYGWGSGEWMDAWKQRTEDTg. ATVIGT-----------AIVNEM--PDNAPE-CKEl. GEAAAKA----hhhhhh eeeee h hhhh STANSIA-GGSDIALLTILNHLMVK-g. MLVYSG----GVAFGKPKTHLG-----YVHINEI--QENEDENARIf. GERi. ANk. V--KQIF-hhhhhhh eeeee hhhhhh h ------TAEAKKENIIAAAQAGASGY-------------VVK----P-FTAATLEEKLNKIFEKLGM-----ess hhhhhtt see ees s hhhhhhhht G Here, the secondary structures for 10 sequences are predicted by the method PREDATOR, while those for the four sequences with 4 -let (PDB) codes are observed in the corresponding PDB tertiary structures
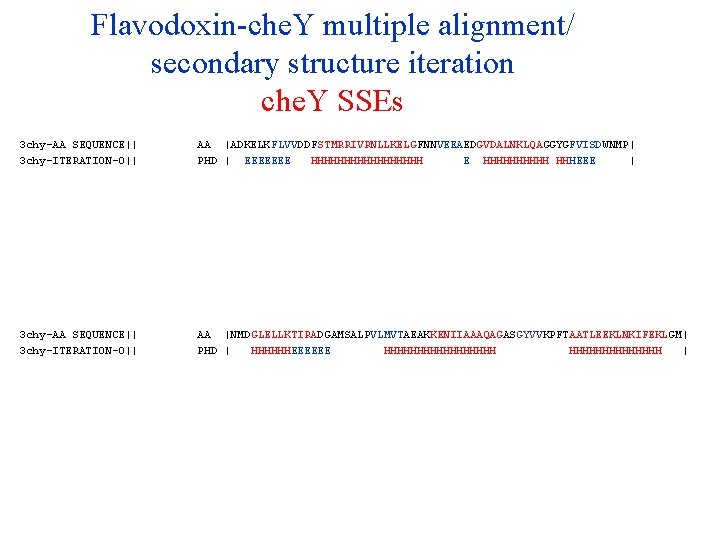
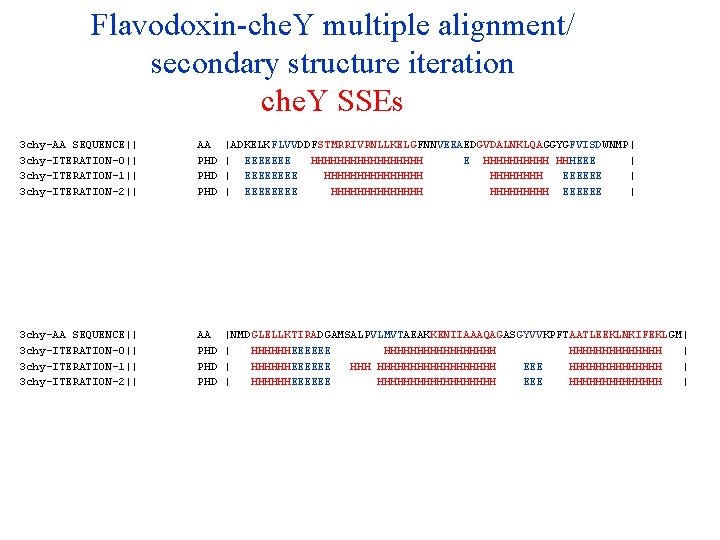
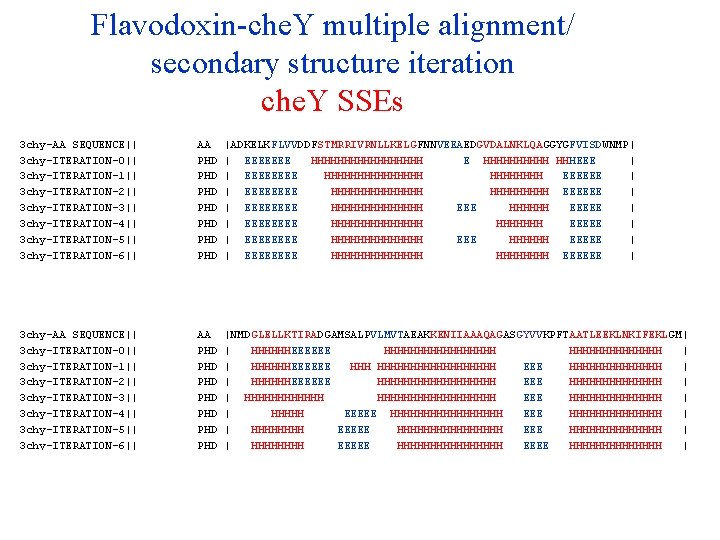
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
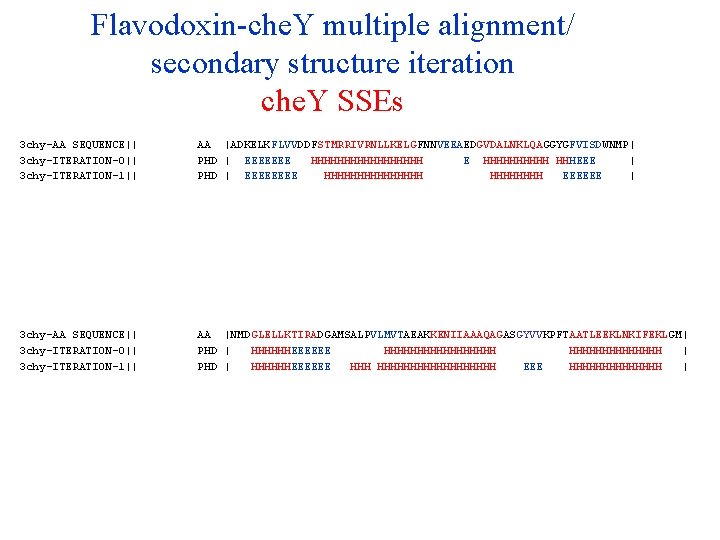
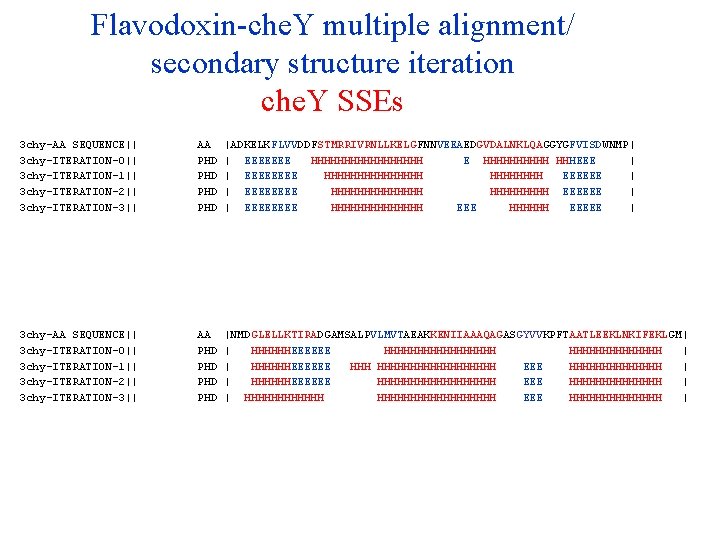
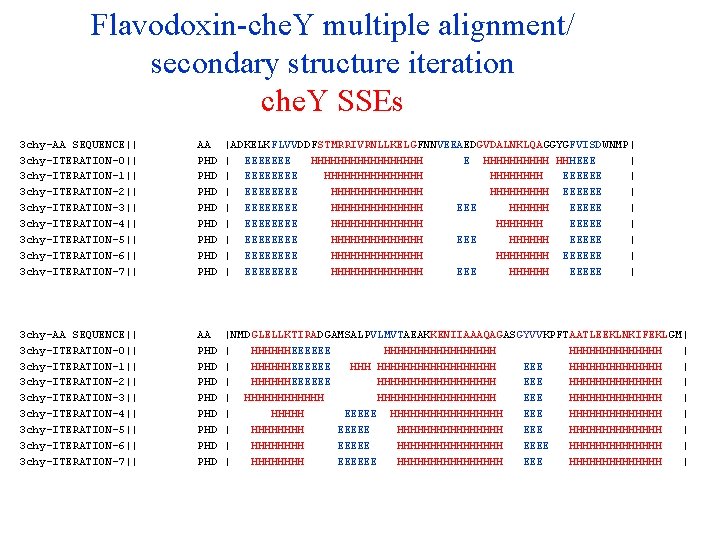
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
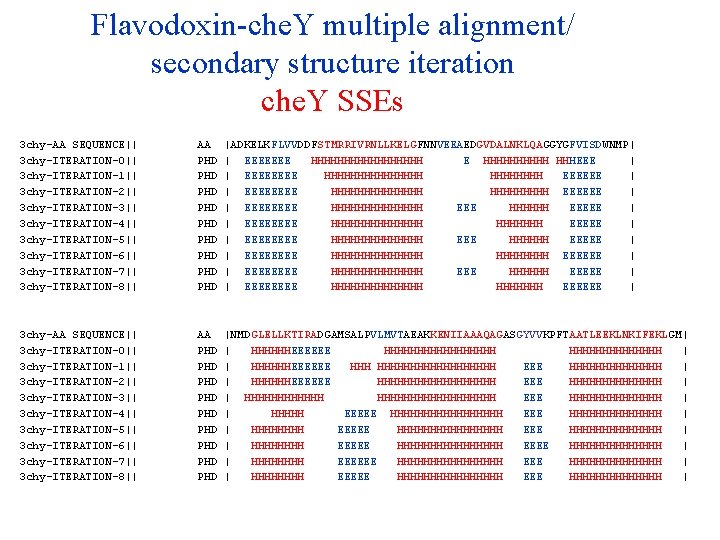
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
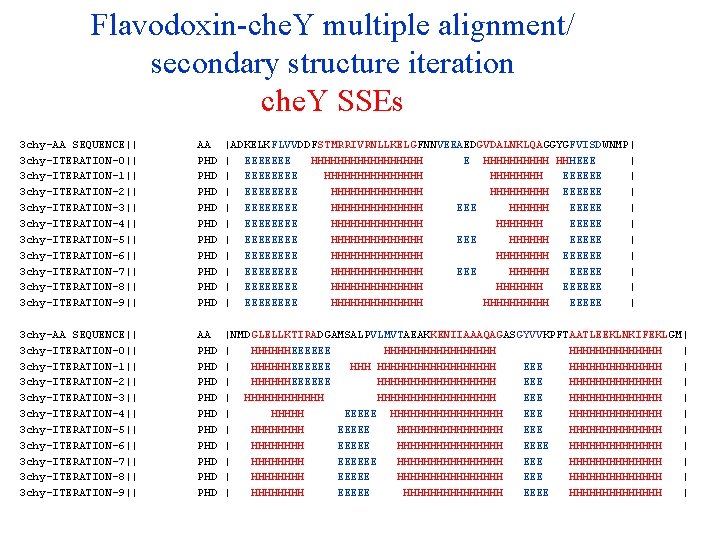
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
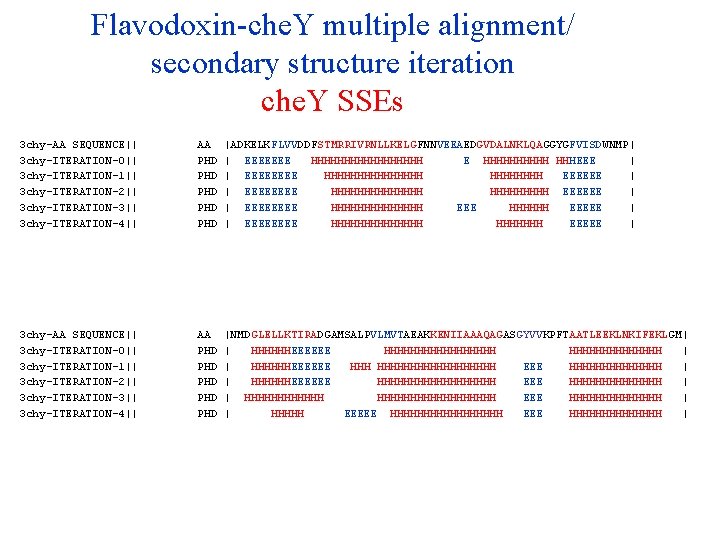
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
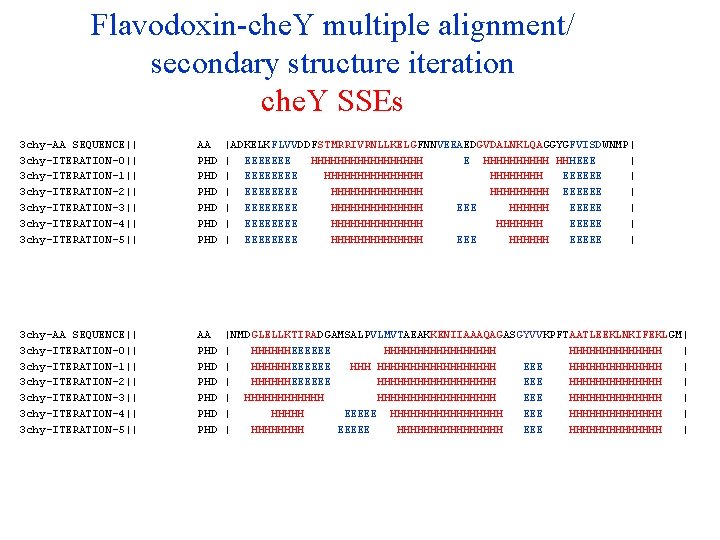
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
Flavodoxin-che. Y multiple alignment/ secondary structure iteration che. Y SSEs 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQAGGYGFVISDWNMP| | EEEEEEE HHHHHHHHH E HHHHH HHHEEE | | EEEE HHHHHHHH EEEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | | EEEE HHHHHHH EEEEEE | | EEEE HHHHHHH EEEEE | 3 chy-AA SEQUENCE|| 3 chy-ITERATION-0|| 3 chy-ITERATION-1|| 3 chy-ITERATION-2|| 3 chy-ITERATION-3|| 3 chy-ITERATION-4|| 3 chy-ITERATION-5|| 3 chy-ITERATION-6|| 3 chy-ITERATION-7|| 3 chy-ITERATION-8|| 3 chy-ITERATION-9|| AA PHD PHD PHD |NMDGLELLKTIRADGAMSALPVLMVTAEAKKENIIAAAQAGASGYVVKPFTAATLEEKLNKIFEKLGM| | HHHHHHEEEEEE HHHHHHHHH | | HHHHHHEEEEEE HHHHHHHHHHH EEE HHHHHHH | | HHHHHHEEEEEE HHHHHHHHH EEE HHHHHHH | | HHHHHHHHHHHHHHHHHH EEE HHHHHHH | | HHHHH EEEEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHHHHHHHHHH EEEE HHHHHHH | | HHHH EEEEEE HHHHHHHHHHHHHHHH EEE HHHHHHHHHHHHHH | | HHHHHHHH EEEEE HHHHHHHH EEEE HHHHHHH |
Iteration Convergence Limit cycle Divergence
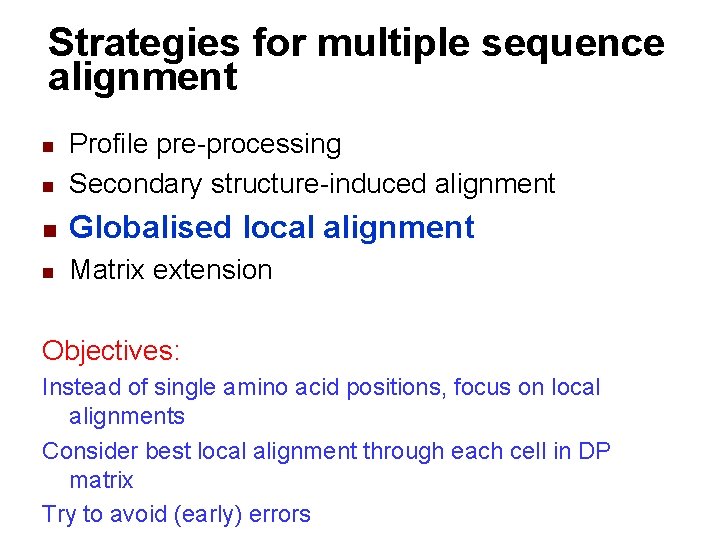
Strategies for multiple sequence alignment n Profile pre-processing Secondary structure-induced alignment n Globalised local alignment n Matrix extension n Objectives: Instead of single amino acid positions, focus on local alignments Consider best local alignment through each cell in DP matrix Try to avoid (early) errors
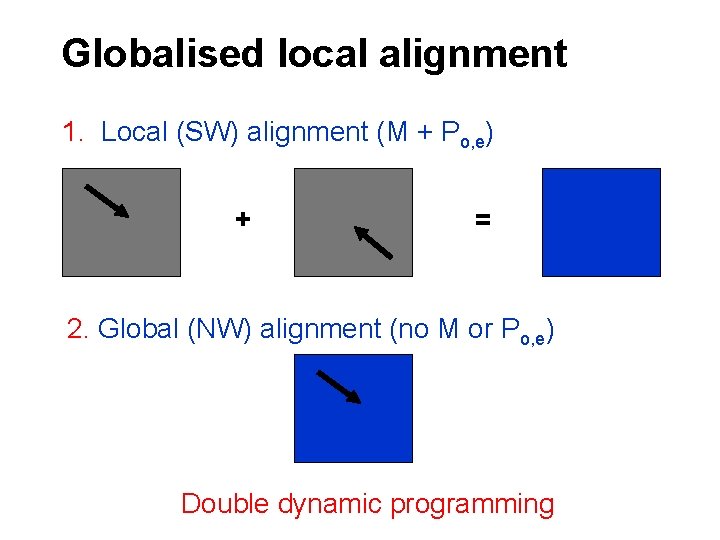
Globalised local alignment 1. Local (SW) alignment (M + Po, e) + = 2. Global (NW) alignment (no M or Po, e) Double dynamic programming
Globalised local alignment 1. 2.
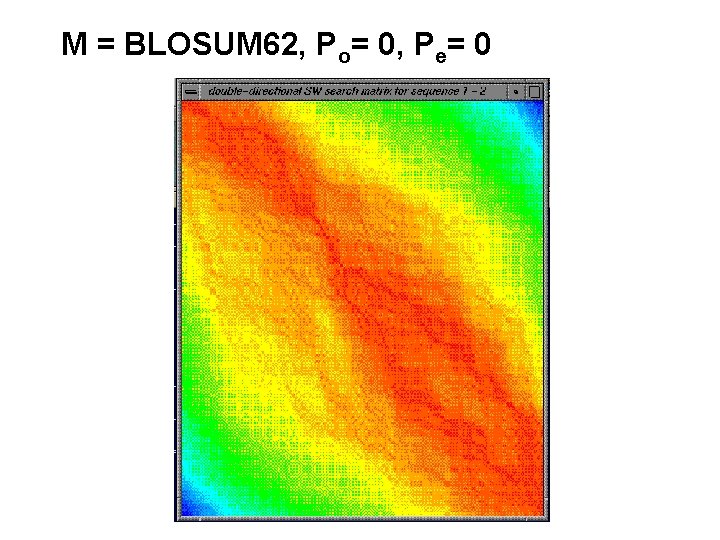
M = BLOSUM 62, Po= 0, Pe= 0
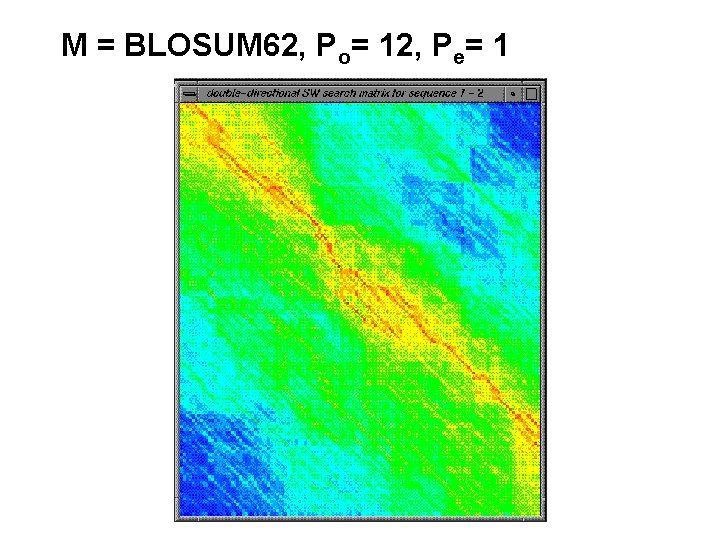
M = BLOSUM 62, Po= 12, Pe= 1
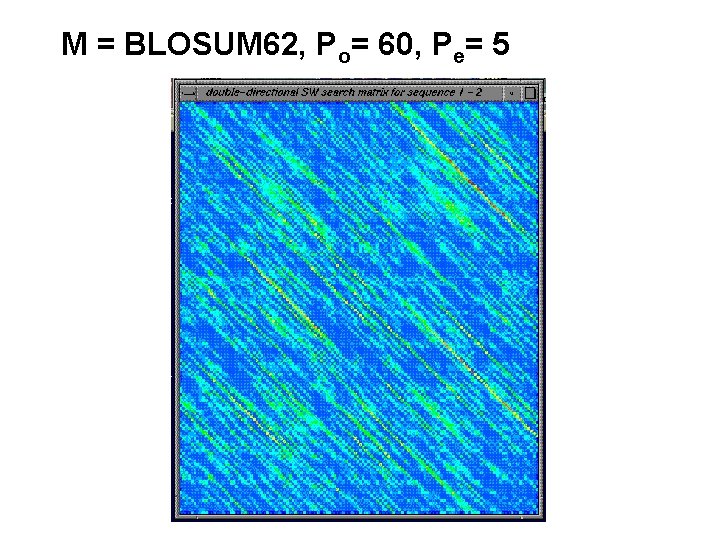
M = BLOSUM 62, Po= 60, Pe= 5
Strategies for multiple sequence alignment n Profile pre-processing Secondary structure-induced alignment Globalised local alignment n Matrix extension n n Objective: try to avoid (early) errors
Integrating alignment methods and alignment information with T -Coffee • Integrating different pair-wise alignment techniques (NW, SW, . . ) • Combining different multiple alignment methods (consensus multiple alignment) • Combining sequence alignment methods with structural alignment techniques • Plug in user knowledge

Matrix extension T-Coffee Tree-based Consistency Objective Function For alignm. Ent Evaluation Cedric Notredame Des Higgins Jaap Heringa J. Mol. Biol. , 302, 205 -217; 2000
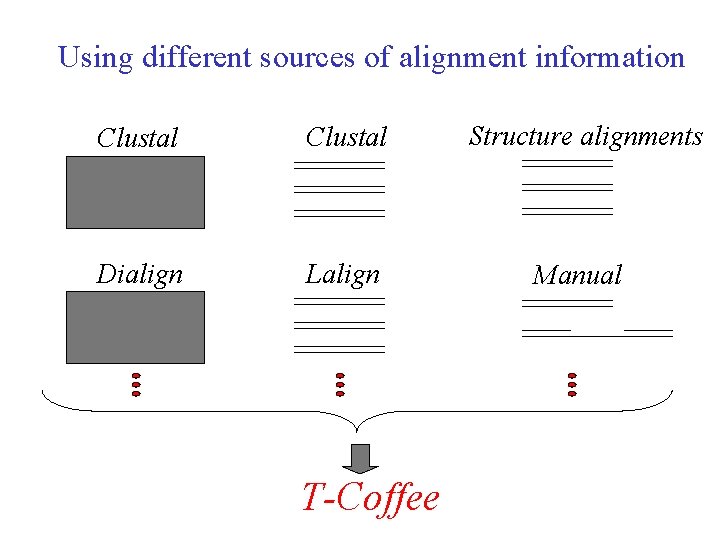
Using different sources of alignment information Clustal Structure alignments Dialign Lalign Manual T-Coffee
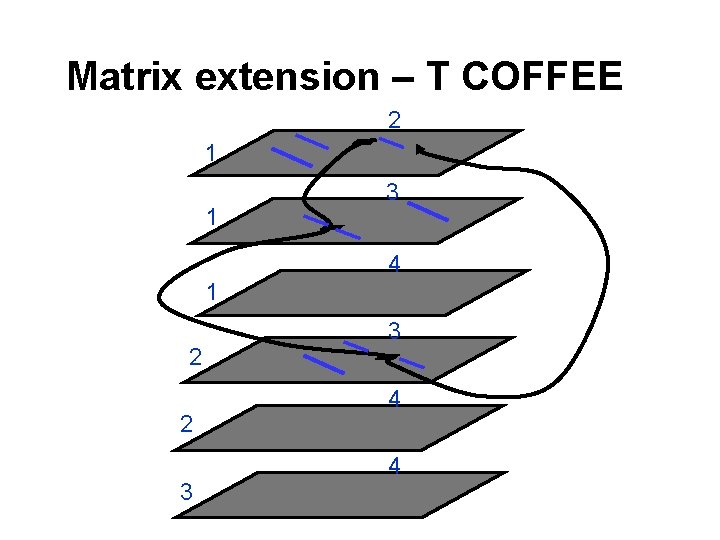
Matrix extension – T COFFEE 2 1 1 3 4 1 2 2 3 3 4 4
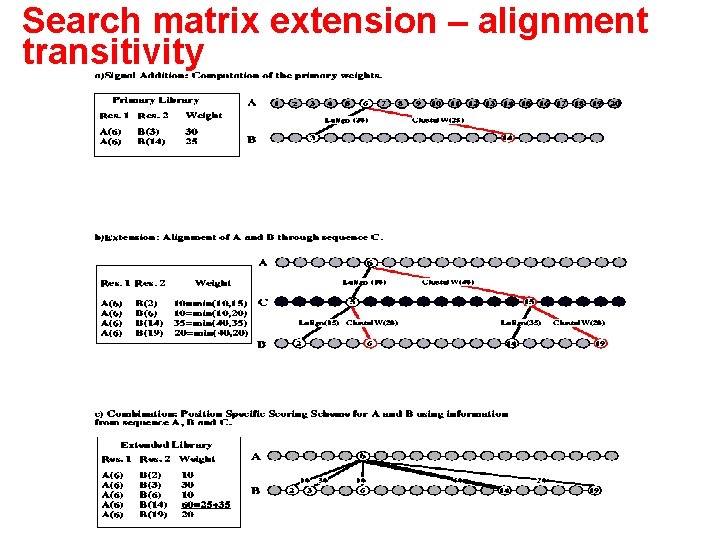
Search matrix extension – alignment transitivity
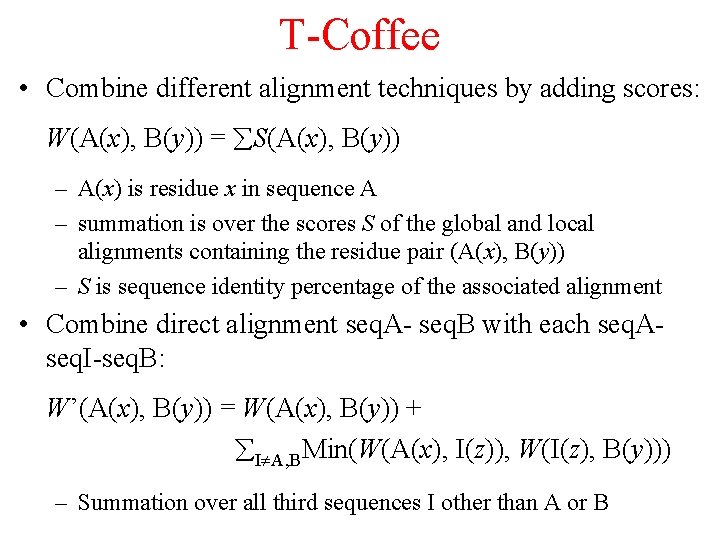
T-Coffee • Combine different alignment techniques by adding scores: W(A(x), B(y)) = S(A(x), B(y)) – A(x) is residue x in sequence A – summation is over the scores S of the global and local alignments containing the residue pair (A(x), B(y)) – S is sequence identity percentage of the associated alignment • Combine direct alignment seq. A- seq. B with each seq. Aseq. I-seq. B: W’(A(x), B(y)) = W(A(x), B(y)) + I A, BMin(W(A(x), I(z)), W(I(z), B(y))) – Summation over all third sequences I other than A or B
T-Coffee Other sequences Direct alignment
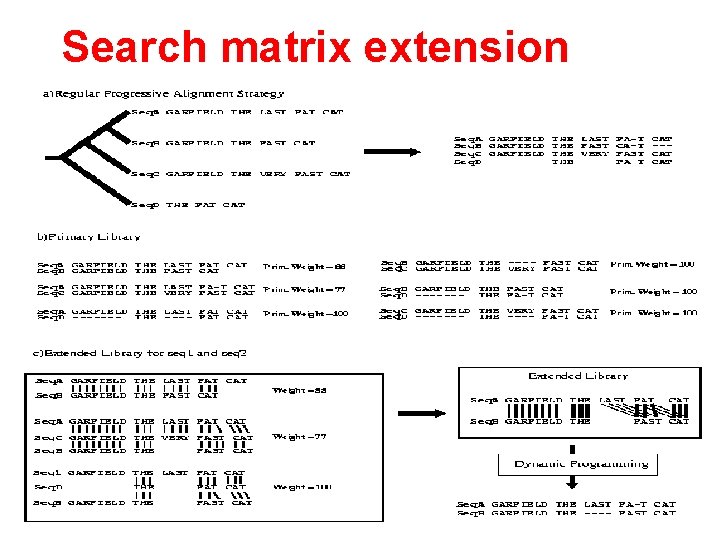
Search matrix extension

Succesful current MSA method: MUSCLE (Edgar, 2004) n n n MUSCLE is very fast and can handle large sets of long sequences MUSCLE features a slightly changed way of profile -profile alignment scoring MUSCLE uses iteration to realign sequences that are together in subgroups (subtrees in the alignment guide tree produced using UPGMA (group averaging - see lecture 4)

Most succesful current MSA method: PSI-PRALINE (Simossis et al. , 2005) n n n PSI-PRALINE uses database searching to find ‘background’ sequences – these are not aligned but aid correct matching of the sequences PSI-PRALINE is slow because it has to do a sequence database search for each sequence PSI-PRALINE is very good at aligning distant sequences
Evaluating multiple alignments n n There are reference databases based on structural information: e. g. BAli. BASE and HOMSTRAD Conflicting standards of truth u u u n n n evolution structure function With orphan sequences no additional information Benchmarks depending on reference alignments Quality issue of available reference alignment databases Different ways to quantify agreement with reference alignment (sum-of-pairs, column score) “Charlie Chaplin” problem
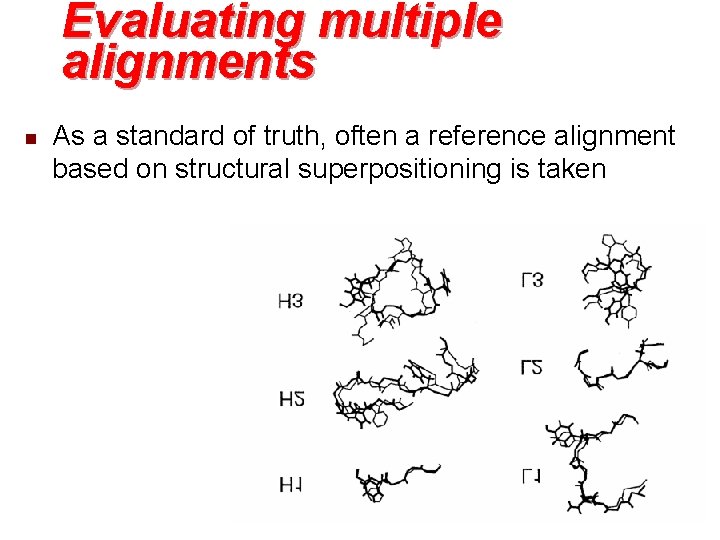
Evaluating multiple alignments n As a standard of truth, often a reference alignment based on structural superpositioning is taken
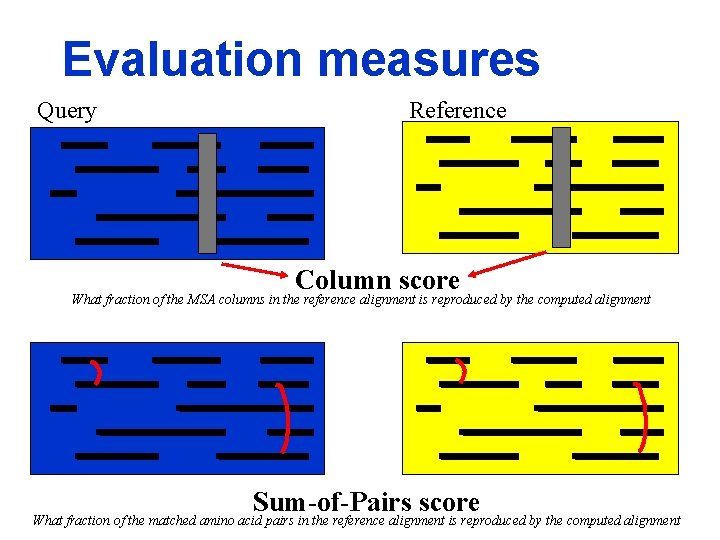
Evaluation measures Query Reference Column score What fraction of the MSA columns in the reference alignment is reproduced by the computed alignment Sum-of-Pairs score What fraction of the matched amino acid pairs in the reference alignment is reproduced by the computed alignment
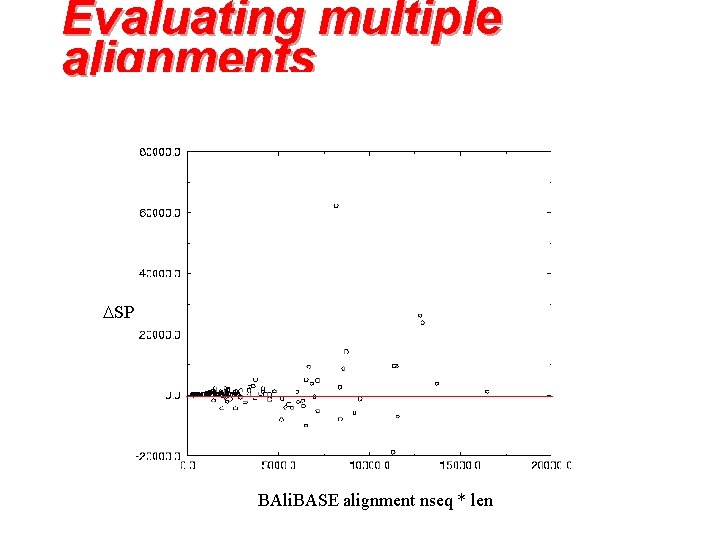
Evaluating multiple alignments SP BAli. BASE alignment nseq * len
Summary n Weighting schemes are developed to minimise (early) errors during the progressive alignment protocol: PRALINE Profile pre-processing (global/local) u T-Coffee Matrix extension (well balanced scheme) u n Smoothing alignment signals u n Using additional information u n PRALINE globalised local alignment PRALINE secondary structure driven alignment Schemes strike balance between speed and sensitivity
References n n Heringa, J. (1999) Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comp. Chem. 23, 341 -364. Notredame, C. , Higgins, D. G. , Heringa, J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. , 302, 205 -217. Heringa, J. (2002) Local weighting schemes for protein multiple sequence alignment. Comput. Chem. , 26(5), 459477. Simossis, V. A. , Kleinjung, J. and Heringa, J. (2005) Homology-extended sequence alignment. Nucleic Acids Res. 33(3): 816 -824.

http: //ibivu. cs. vu. nl/teaching/mnw 2_2005. php
- Slides: 47