Introduction to bioinformatics lecture 9 Multiple sequence alignment
Introduction to bioinformatics lecture 9 Multiple sequence alignment (II)
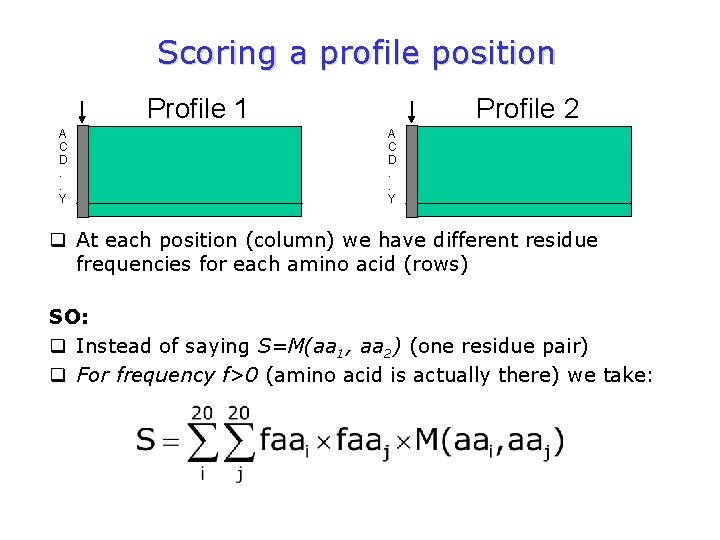
Scoring a profile position Profile 1 A C D. . Y Profile 2 A C D. . Y q At each position (column) we have different residue frequencies for each amino acid (rows) SO: q Instead of saying S=M(aa 1, aa 2) (one residue pair) q For frequency f>0 (amino acid is actually there) we take:
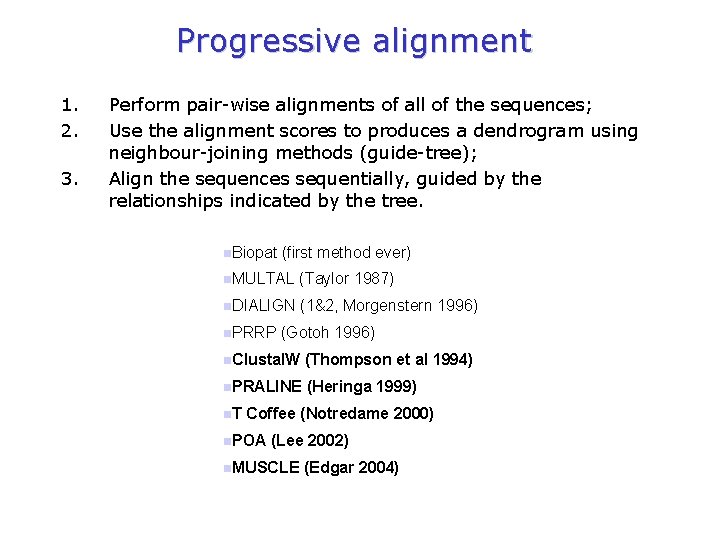
Progressive alignment 1. 2. 3. Perform pair-wise alignments of all of the sequences; Use the alignment scores to produces a dendrogram using neighbour-joining methods (guide-tree); Align the sequences sequentially, guided by the relationships indicated by the tree. n. Biopat (first method ever) n. MULTAL (Taylor 1987) n. DIALIGN (1&2, Morgenstern 1996) n. PRRP (Gotoh 1996) n. Clustal. W (Thompson et al 1994) n. PRALINE (Heringa 1999) n. T Coffee (Notredame 2000) n. POA (Lee 2002) n. MUSCLE (Edgar 2004)
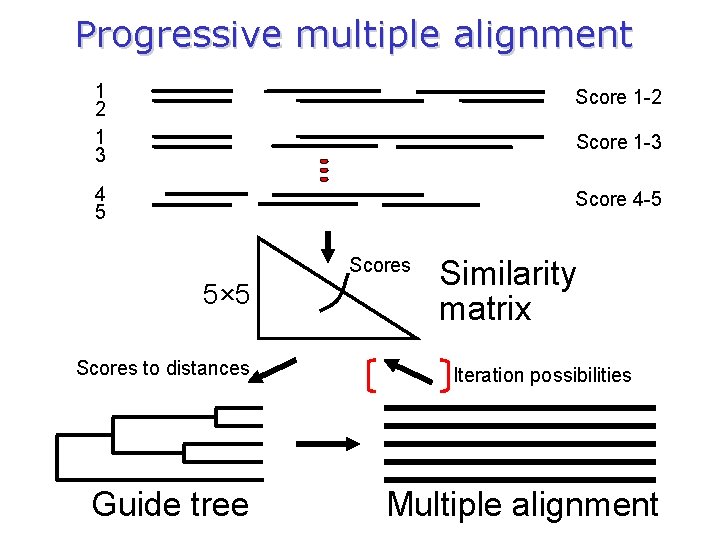
Progressive multiple alignment 1 2 1 3 Score 1 -2 4 5 Score 4 -5 Score 1 -3 Scores 5× 5 Scores to distances Guide tree Similarity matrix Iteration possibilities Multiple alignment
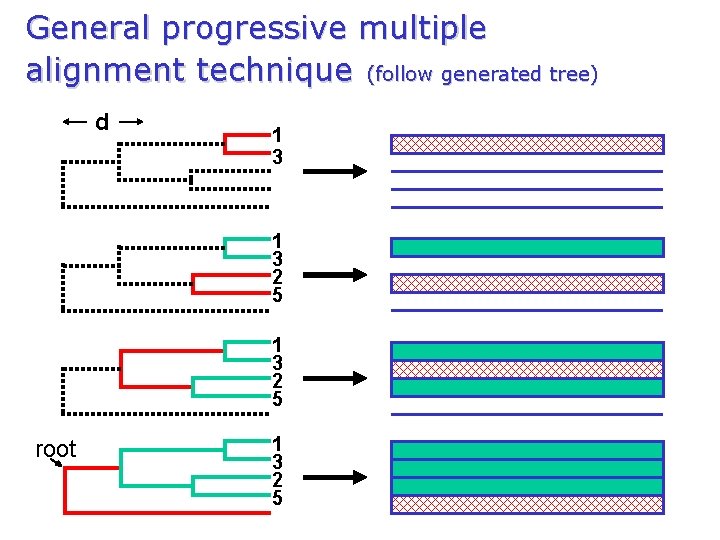
General progressive multiple alignment technique (follow generated tree) d 1 3 2 5 root 1 3 2 5
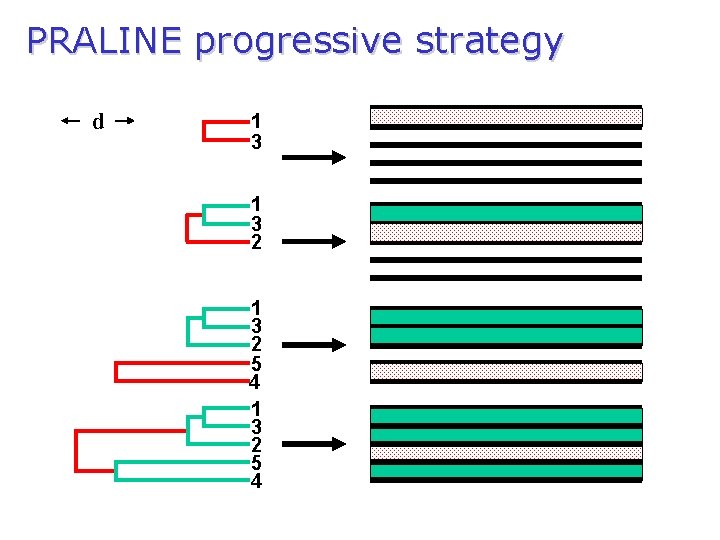
PRALINE progressive strategy d 1 3 2 5 4

There are problems … Accuracy is very important !!!! q Alignment errors during the construction of the MSA cannot be repaired anymore: propagated into the progressive steps. q The comparisons of sequences at early steps during progressive alignments cannot make use of information from other sequences. q It is only later during the alignment progression that more information from other sequences (e. g. through profile representation) becomes employed in the alignment steps. “Once a gap, always a gap” Feng & Doolittle, 1987

Additional strategies for multiple sequence alignment • Profile pre-processing • Secondary structure-induced alignment • Globalised local alignment • Matrix extension Objective: try to avoid (early) errors
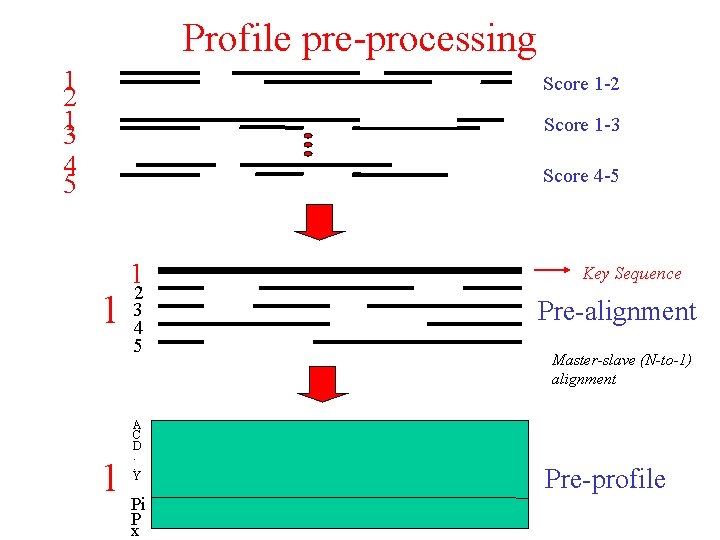
Profile pre-processing 1 2 13 Score 1 -2 Score 1 -3 4 5 Score 4 -5 1 1 2 3 4 5 A C D. . Y 1 Pi P x Key Sequence Pre-alignment Master-slave (N-to-1) alignment Pre-profile
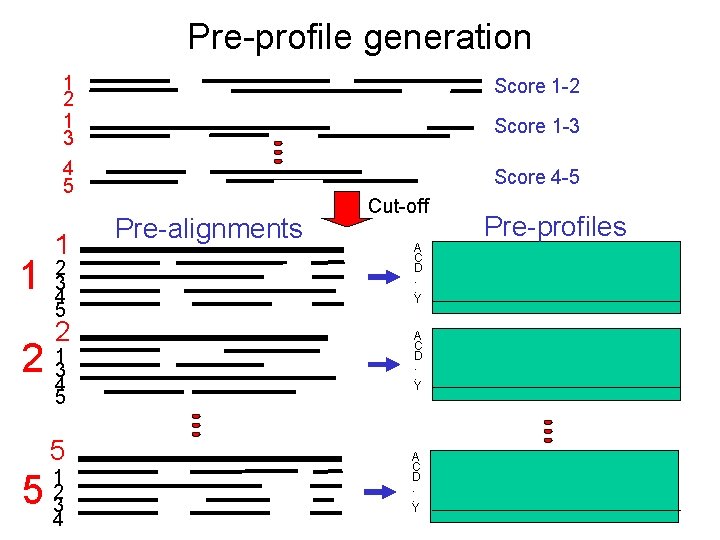
Pre-profile generation 1 1 2 1 3 Score 1 -2 4 5 Score 4 -5 1 2 3 4 5 2 2 134 5 5 5 1 2 3 4 Score 1 -3 Pre-alignments Cut-off A C D. . Y Pre-profiles
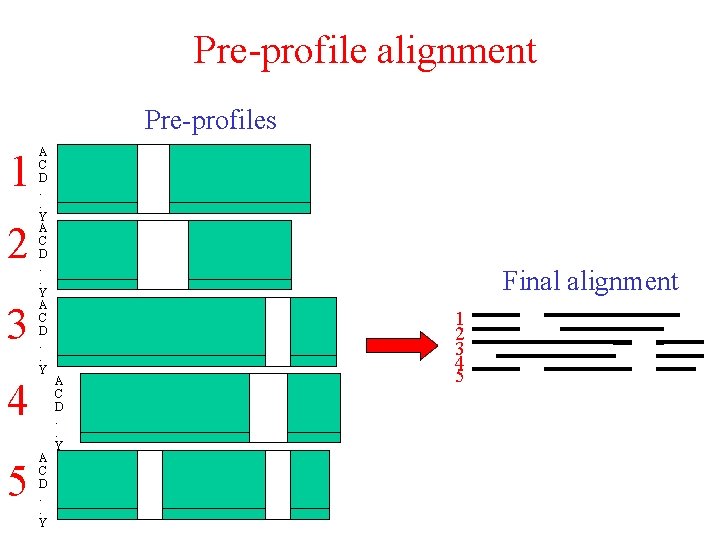
Pre-profile alignment Pre-profiles 1 2 3 4 5 A C D. . Y Final alignment A C D. . Y 1 2 3 4 5
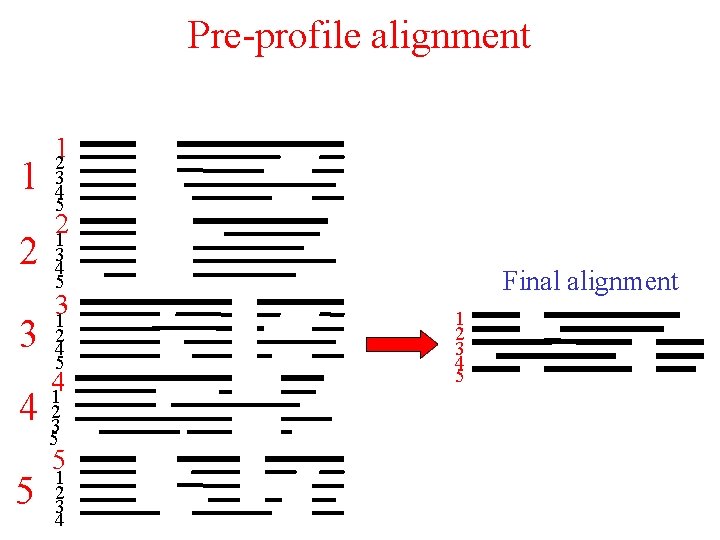
Pre-profile alignment 1 2 3 12 3 4 5 21 3 4 5 31 2 4 5 41 4 23 5 5 5 1 2 3 4 Final alignment 1 2 3 4 5
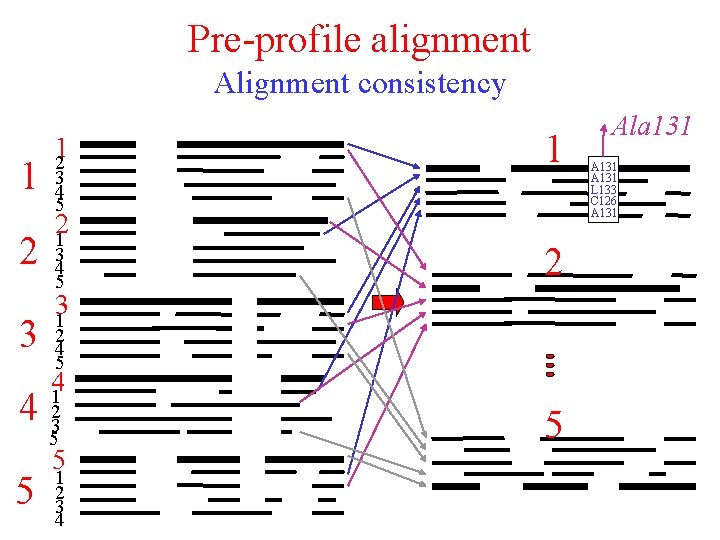
Pre-profile alignment Alignment consistency 1 2 3 12 3 4 5 21 3 4 5 2 31 2 4 5 41 4 23 5 5 1 2 3 4 5 Ala 131 A 131 L 133 C 126 A 131

PRALINE pre-profile generation • Idea: use the information from all query sequences to make a pre-profile for each query sequence that contains information from other sequences • You can use all sequences in each pre-profile, or use only those sequences that will probably align ‘correctly’. Incorrectly aligned sequences in the preprofiles will increase the noise level. • Select using alignment score: only allow sequences in pre-profiles if their alignment with the score higher than a given threshold value. In PRALINE, this threshold is given as prepro=1500 (alignment score threshold value is 1500 – see next two slides)

Flavodoxin-che. Y consistency scores (PRALINE prepro=0) 1 fx 1 FLAV_DESVH FLAV_DESDE FLAV_DESGI FLAV_DESSA 4 fxn FLAV_MEGEL 2 fcr FLAV_ANASP FLAV_ECOLI FLAV_AZOVI FLAV_ENTAG FLAV_CLOAB 3 chy --78999999 TEYTAETIARQL 8776 -6657777777553799 VL 999 ST 97775599989 -435566677798998878 AQGRKVACF -4678899999 TEYTAETIAREL 7777 -7757777777553799 VL 999 ST 97775599989 -435566677798998878 AQGRKVACF -478999999999998877669565888877778763 YDAVL 999 SAW 9877789877753556666669777776789 GRKVAAF -4678899999 TEGVAEAIAKTL 9997 -76678888777777887539 DVVL 999 ST 987776 --9889546667776697776557777888888 9367779999999999988759765777888887639999 STW 77765 --99995366666777979987799999 -8787799999999997766669675677888888777999999988777776 --988957778888889777323788888 9776779999999997777766 -66566667778889997679999987777669 --887362334466695555455778888888 --878999999 TEVADFIGK 996541900300000112233355679 DLLF 9999985531288811122455555540777777788888 -47899 LFYGTQTGKTESVAEIIR 9777653922356677778977799999988843 --99985557787778999988799999 997789999 GSDTGNTENIAKMIQ 87742229224566788899999955699999755553 ----99262225555495777767778999999 --79 IGLFFGSNTGKTRKVAKSIK 99887759657577888888999777899999987776111222244555 -5555555778999999 947899999999999875522922323455555568889999999887552111133477777 -777777799999 -86999 ILYSSKTGKTERVAK 999755555505767888887777765778899998522223 --98883422344555977777777 0122222223333335666665555555222922222221112163335555755553222888877674533344493332222222 Avrg Consist Conservation 86677788888999998776554844455566666555788888766544887666334445566586666556778888888 0125538675848969746963946463343045244355446543473516658868567554455000000314365446505575435547747759 1 fx 1 FLAV_DESVH FLAV_DESDE FLAV_DESGI FLAV_DESSA 4 fxn FLAV_MEGEL 2 fcr FLAV_ANASP FLAV_ECOLI FLAV_AZOVI FLAV_ENTAG FLAV_CLOAB 3 chy G 888799955555559888888888899777 ----7777797787787978 ---555555566776555677777778888799 -----A 88878685555555999988888889998879 --8777788 -98777777 --8555544332456677777599 -----877759777555556777777778 ---888888876677787777755555542424667888887777 -------977768777555556777777776788777778888 -978985555565365568888877 -------8677775555266666555555577887767999877777977777665555544446666555798 -----857777566666652555677777888888868997788898877655867788554433322222221223355557 -------877773573333333777766667777765533333333228333332244444567777777888777633 -----9777737753333447778888887777777333344444433833333344444455577777788777734 -----97774378644444477778888883333444444424444455555455577566778888877734110000 977763553333334666666677777333344444448233335555554555888877772311 ---97777388655555586666677666633333333221233333444445555566666555582 -----76662722222221244444555555878822222221111111222222344443333333233399 -----22222722224111355431113324578 -877789976665568777763222222322222323344444422 ------ Avrg Consist Conservation 866656564444444666666665666555555565444443344455666666889999 73663057433334163464534444*746710000011010011000000010434744645443225474454448434301000000 Consistency are scored 0 to. SId= 10; 3838 the Av. SId= value 0. 297 10 is represented by the corresponding amino acid (red) Iteration 0 values SP= 135136. 00 Av. SP=from 10. 473
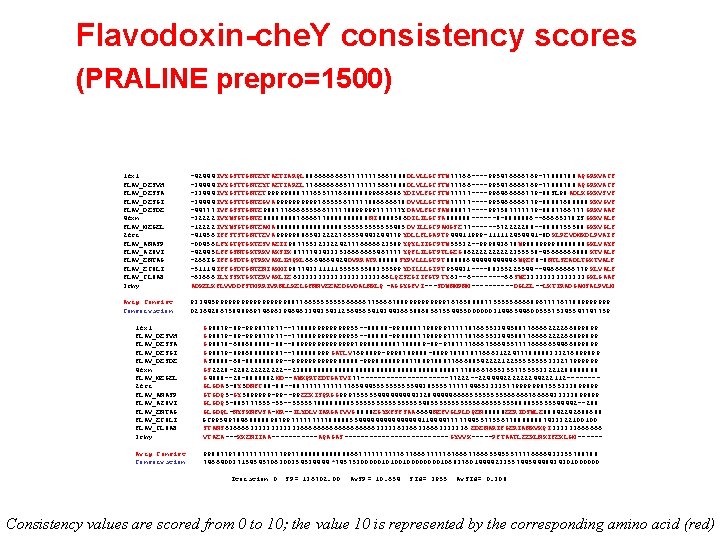
Flavodoxin-che. Y consistency scores (PRALINE prepro=1500) 1 fx 1 FLAV_DESVH FLAV_DESSA FLAV_DESGI FLAV_DESDE 4 fxn FLAV_MEGEL 2 fcr FLAV_ANASP FLAV_AZOVI FLAV_ENTAG FLAV_ECOLI FLAV_CLOAB 3 chy -42444 IVYGSTTGNTEYTAETIARQL 886666666577777775667888 DLVLLGCSTW 77766 ----995476666769 -77888788 AQGRKVACF -34444 IVYGSTTGNTEYTAETIAREL 776666666577777775667888 DLVLLGCSTW 77766 ----995476666769 -77888788 AQGRKVACF -33444 IVYGSTTGNTET 99999888777655777668888899666686 YDIVLFGCSTW 77777 ----996466666779 -88 SL 98 ADLKGKKVSVF -34444 IVYGSTTGNTEGVA 99999765555677777886666678 DVVLLGCSTW 77777 ----995466666779 -88887688888 KKVGVF -44777 IVFGSSTGNTE 988777666655566777778899999777777 Y DAVLFGCSAW 88877 ----997587777779 -8887766777 GRKVAAF -32222 IVYWSGTGNTE 8888766667788888 NI 8888586 DILILGCSA 888888 ------8 -8888886 --66665378 IS GKKVALF -12222 IVYWSGTGNTEAMA 88888888555555485 DVILLGCPAMGSE 77 ------572222288 --8888755588 GKKVGLF -41456 IFFSTSTGNTTEVA 999998865432222765554443244779 YDLLFLGAPT 944411999 -111112454441 -8 D KLPEVDMKDLPVAIF -00456 LFYGTQTGKTESVAEII 987755323322427776666623589 YQYLIIGCPTW 55532 --999843678 W 988899998888888 GKLVAYF -42445 LFFGSNTGKTRKVAKSIK 87777434333536666665467777 YQFLILGTPTLGEG 8622222355558 -45666666888 KTVALF -266 IG IFFGSDTGQTRKVAKLIHQKL 6664664424 DVRRATR 88888 SYPVLLLGTPT 888886444446 WQEF 8 -8 NTLSEADLTGKTVALF -51114 IFFGSDTGNTENIAKMI 987743311111555555588355599 YDILLLGIPT 954431 ----88355225544 --44666666779 KLVALF -63666 ILYSSKTGKTERVAKLIE 6333333333366 LQESEGIIFGTPTY 63 --6 ----66 SWE 3333333 GKLGAAF ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQ -AGGYGFVI---SDWNMPNM-----DGLEL--LKTIRADGAMSALPVLM Avrg Consist Conservation 93344599999999988776655556666677566678899999767658888775555566668967777677889999999 0236428675848969746963946463344354312564565414344366588685675544550000003144654460055575345547747759 1 fx 1 FLAV_DESVH FLAV_DESSA FLAV_DESGI FLAV_DESDE 4 fxn FLAV_MEGEL 2 fcr FLAV_ANASP FLAV_AZOVI FLAV_ENTAG FLAV_ECOLI FLAV_CLOAB 3 chy G 98879 -89 -999877977 --7788899999999955 --88888 -9988887798999777778766553344588776666222266899899 G 98878 -688688888 -88 --88999999979988888887788889 -89 -9787777666756645577776666654466899899 G 98879 -898688888987 --788888999 GATLV 7698899 -9998789888 -8899787878776663122477788888333276899899 AS 8888 -68 -888888899 --99999988888 -9998888898877889788877666885422221225555333277999999 GS 2228 -22822222 --238888888888888888888777886676553557755553322128888 G 4888 --28 -8888882 MD--AWKQRTEDTGATVI 77 -----------77222 --224444222222244222112 -------GLGDA 5 -8 Y 5 DNFC 88 -88 --88777777654445555544385555777774465333357799999987555333899899 GTGDQ 5 -GY 5899999 -99 --99 EEKISQRGG 999755555444443328444446666555556666676666433333899899 GLGDQ 5 -885777555 -55 --555557888855555555548555555666555555888855555544442 --288 GLGDQL-NYSKNFVSA-MR--ILYDLVIARGACVVG 8888 EGYKFSFSAA 6664 NEFVGLPLDQEN 88888 EERIDSWLE 88842242688688 GC 995497846888889879977777888885544444444114444777774455775567788888887433322100100 STANS 636666333333666666666333336336663333336 EDENARIFGERIANKVKQI 333333666666 VTAEA---KKENIIAA------AQAGAS------------- GYVVK-----PFTAATLEEKLNKIFEKLGM ------ Avrg Consist Conservation 998877978777779977888888667777767766677777676667766655455577776666433355788788 746640037154545706300354534444 *74575300000101001000010683760144442335574454448434301000000 Iteration 0 SP= 136702. 00 Av. SP= 10. 654 SId= 3955 Av. SId= 0. 308 Consistency values are scored from 0 to 10; the value 10 is represented by the corresponding amino acid (red)
Strategies for multiple sequence alignment • Profile pre-processing • Secondary structure-induced alignment • Globalised local alignment • Matrix extension Objective: integrate secondary structure information to anchor alignments and avoid errors
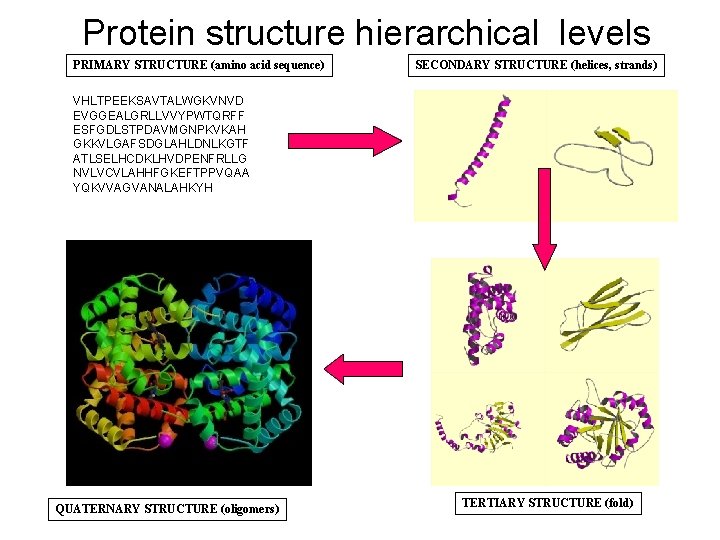
Protein structure hierarchical levels PRIMARY STRUCTURE (amino acid sequence) SECONDARY STRUCTURE (helices, strands) VHLTPEEKSAVTALWGKVNVD EVGGEALGRLLVVYPWTQRFF ESFGDLSTPDAVMGNPKVKAH GKKVLGAFSDGLAHLDNLKGTF ATLSELHCDKLHVDPENFRLLG NVLVCVLAHHFGKEFTPPVQAA YQKVVAGVANALAHKYH QUATERNARY STRUCTURE (oligomers) TERTIARY STRUCTURE (fold)
Why use (predicted) structural information • “Structure more conserved than sequence” – Many structural protein families (e. g. globins) have family members with very low sequence similarities. For example, globin sequences identities can be as low as 10% while still having an identical fold. • This means that you can still observe equivalent secondary structures in homologous proteins even if sequence similarities are extremely low. • But you are dependent on the quality of prediction methods. For example, secondary structure prediction is currently at 76% correctness. So, 1 out of 4 predicted amino acids is still incorrect.
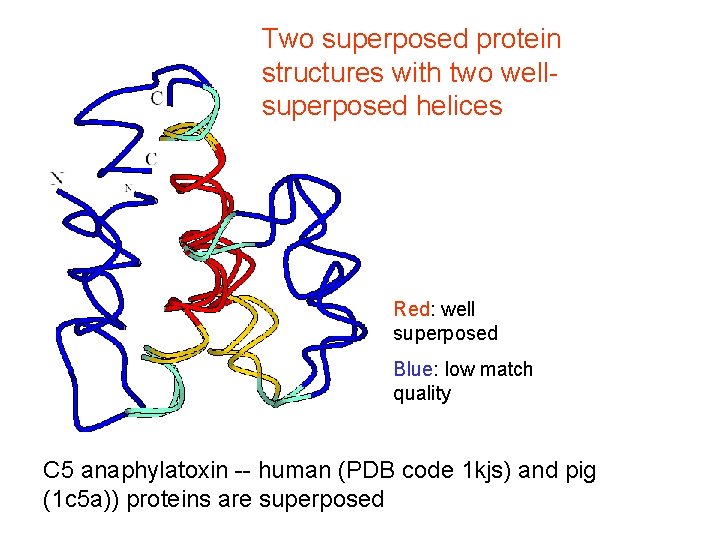
Two superposed protein structures with two wellsuperposed helices Red: well superposed Blue: low match quality C 5 anaphylatoxin -- human (PDB code 1 kjs) and pig (1 c 5 a)) proteins are superposed
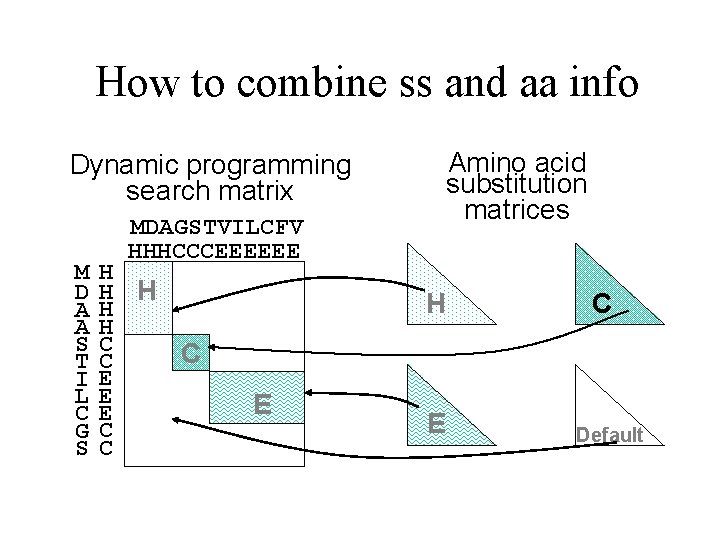
How to combine ss and aa info Amino acid substitution matrices Dynamic programming search matrix M D A A S T I L C G S MDAGSTVILCFV HHHCCCEEEEEE H H H C C C E E E C C H E E C Default

In terms of scoring… • So how would you score a profile using this extra information? – Same formula as in lecture 6, but you can use sec. struct. specific substitution scores in various combinations. • Where does it fit in? – Very important: structure is always more conserved than sequence so it can help with the insertion(or not) of gaps.
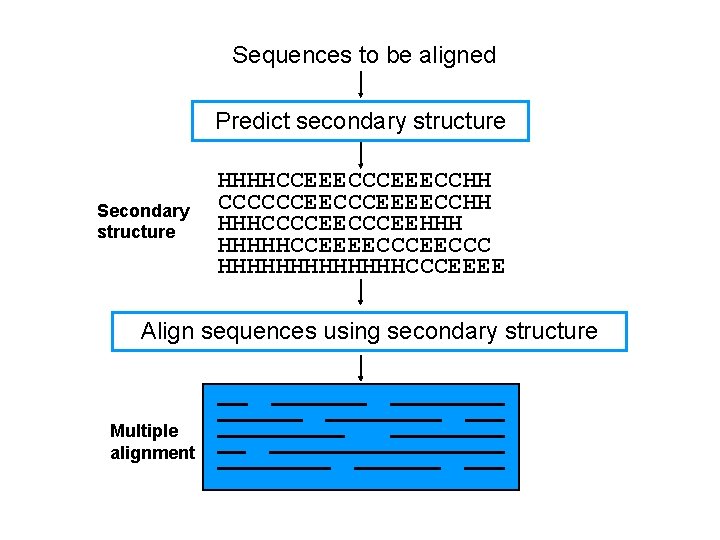
Sequences to be aligned Predict secondary structure Secondary structure HHHHCCEEECCHH CCCCCCEEEECCHH HHHCCCCEEHHH HHHHHCCEEEECCC HHHHHHHCCCEEEE Align sequences using secondary structure Multiple alignment
Using predicted secondary structure 1 fx 1 FLAV_DESVH FLAV_DESGI FLAV_DESSA FLAV_DESDE 2 fcr FLAV_ANASP FLAV_ECOLI FLAV_AZOVI FLAV_ENTAG 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy -PK-ALIVYGSTTGNTEYTAETIARQLANAG-YEVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSI------ELQDDFIPLFDS-LEETGAQGRKVACF e eeee b ssshhhhhhhttt eeeee stt tttttt seeee b ee sss ee ttthhhhtt ttss tt eeeee MPK-ALIVYGSTTGNTEYTa. ETIARELADAG-YEVDSRDAASVEAGGLFEGFDLVLLg. CSTWGDDSI------ELQDDFIPLFDS-LEETGAQGRKVACf e eeeeee hhhhhhhh eeeeee hhhhhh eeeee MPK-ALIVYGSTTGNTEGVa. EAIAKTLNSEG-METTVVNVADVTAPGLAEGYDVVLLg. CSTWGDDEI------ELQEDFVPLYED-LDRAGLKDKKVGVf e eeeeee hhhhhhh eeeeee MSK-SLIVYGSTTGNTETAa. EYVAEAFENKE-IDVELKNVTDVSVADLGNGYDIVLFg. CSTWGEEEI------ELQDDFIPLYDS-LENADLKGKKVSVf eeeeee hhhhhhh eeeee MSK-VLIVFGSSTGNTESIa. QKLEELIAAGG-HEVTLLNAADASAENLADGYDAVLFg. CSAWGMEDL------EMQDDFLSLFEE-FNRFGLAGRKVAAf eeee hhhhhhh eeeee hhhhhheeeee hhhhh eeeee --K-IGIFFSTSTGNTTEVADFIGKTLGAK---ADAPIDVDDVTDPQALKDYDLLFLGAPTWNTGAD----TERSGTSWDEFLYDKLPEVDMKDLPVAIF eeeee ssshhhhhhhggg b eeggg s gggggg seeeeeee stt s sthhhhhhhtggg tt eeeee SKK-IGLFYGTQTGKTESVa. EIIRDEFGND--VVTL-HDVSQAE-VTDLNDYQYLIIg. CPTWNIGEL----QSDWEGLYSE-LDDVDFNGKLVAYf eeeee hhhhhhheeeeee hhhhh eeeeee -AI-TGIFFGSDTGNTENIa. KMIQKQLGKD--VADV-HDIAKSS-KEDLEAYDILLLg. IPTWYYGEA----QCDWDDFFPT-LEEIDFNGKLVALf eee hhhhhhheeeee hhhhh eeeeee -AK-IGLFFGSNTGKTRKVa. KSIKKRFDDET-MSDA-LNVNRVS-AEDFAQYQFLILg. TPTLGEGELPGLSSDCENESWEEFLPK-IEGLDFSGKTVALf eee hhhhhhheeeee hhhhh eeeeee MAT-IGIFFGSDTGQTRKVa. KLIHQKLDG---IADAPLDVRRAT-REQFLSYPVLLLg. TPTLGDGELPGVEAGSQYDSWQEFTNT-LSEADLTGKTVALf eeee hhhhhhheeeee hhhhh eeeee ----MKIVYWSGTGNTEKMAELIAKGIIESG-KDVNTINVSDVNIDELLNE-DILILGCSAMGDEVL------E-ESEFEPFIEE-IST-KISGKKVALF eeeee ssshhhhhhhhtt eeeettt sttttt seeeeee btttb ttthhhhhhh hst t tt eeeee M---VEIVYWSGTGNTEAMa. NEIEAAVKAAG-ADVESVRFEDTNVDDVASK-DVILLg. CPAMGSEEL------E-DSVVEPFFTD-LAP-KLKGKKVGLf hhhhhhh eeeee M-K-ISILYSSKTGKTERVa. KLIEEGVKRSGNIEVKTMNL-DAVDKKFLQESEGIIFg. TPTY-YANI----SWEMKKWIDE-SSEFNLEGKLGAAf eee hhhhhhh eeeeee hhhhh eeeee ADKELKFLVVDDFSTMRRIVRNLLKELGFNN-VEEAEDGV-DALNKLQAGGYGFVISD---WNMPNM-----DGLELLKTIRADGAMSALPVLMV tt eeee s hhhhhhht eeeesshh hhhh eeeee s sss hhhhh ttttt eeee GCGDS-SY-EYFCGAVDAIEEKLKNLGAEIVQD-----------GLRIDGD--PRAARDDIVGWAHDVRGAI-------eee s ss sstthhhhhhttt ee s eeees gggghhhhhhh GCGDS-SY-EYFCGAVDAIEEKLKNLg. AEIVQD-----------GLRIDGD--PRAARDDIVGw. AHDVRGAI-------eee hhhhhh eeeee hhhhhhh GCGDS-SY-TYFCGAVDVIEKKAEELg. ATLVAS-----------SLKIDGE--P--DSAEVLDw. AREVLARV-------eee hhhhhh eeeee hhhhhh GCGDS-DY-TYFCGAVDAIEEKLEKMg. AVVIGD-----------SLKIDGD--P--ERDEIVSw. GSGIADKI-------hhhhhh eeeee e eee ASGDQ-EY-EHFCGAVPAIEERAKELg. ATIIAE-----------GLKMEGD--ASNDPEAVASf. AEDVLKQL-------e hhhhhhh eeeee ee hhhhhh GLGDAEGYPDNFCDAIEEIHDCFAKQGAKPVGFSNPDDYDYEESKSVRD-GKFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-----eee ttt ttsttthhhhhhtt eee b gggs s tteet teesseeeettt ss hhhhhhhht GTGDQIGYADNFQDAIGILEEKISQRg. GKTVGYWSTDGYDFNDSKALR-NGKFVGLALDEDNQSDLTDDRIKSw. VAQLKSEFGL-----hhhhhhh eeee hhhhhhhh GCGDQEDYAEYFCDALGTIRDIIEPRg. ATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKw. VKQISEELHLDEILNA hhhhhhh eeee hhhhhhhhh GLGDQVGYPENYLDALGELYSFFKDRg. AKIVGSWSTDGYEFESSEAVVD-GKFVGLALDLDNQSGKTDERVAAw. LAQIAPEFGLS--L-e hhhhhhh eeeee hhhhhh GLGDQLNYSKNFVSAMRILYDLVIARg. ACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSw. LEKLKPAV-L-----hhhhhhhh eeee hhhhhhhhhhhh G-----SYGWGDGKWMRDFEERMNGYGCVVVET-----------PLIVQNE--PDEAEQDCIEFGKKIANI----e eesss shhhhhhtt ee s eeees ggghhhhhht G-----SYGWGSGEWMDAWKQRTEDTg. ATVIGT-----------AIVNEM--PDNAPE-CKEl. GEAAAKA----hhhhhh eeeee h hhhh STANSIA-GGSDIALLTILNHLMVK-g. MLVYSG----GVAFGKPKTHLG-----YVHINEI--QENEDENARIf. GERi. ANk. V--KQIF-hhhhhhh eeeee hhhhhh h ------TAEAKKENIIAAAQAGASGY-------------VVK----P-FTAATLEEKLNKIFEKLGM-----ess hhhhhtt see ees s hhhhhhhht G
Strategies for multiple sequence alignment • Profile pre-processing • Secondary structure-induced alignment • Globalised local alignment • Matrix extension Objectives: Instead of single amino acid positions, focus on local alignments Consider best local alignment through each cell in DP matrix Try to avoid (early) errors
Globalised local alignment 1. Local (SW) alignment (M + Po, e) + = 2. Global (NW) alignment (no M or Po, e) Double dynamic programming
Strategies for multiple sequence alignment • Profile pre-processing • Secondary structure-induced alignment • Globalised local alignment • Matrix extension Objective: try to avoid (early) errors
Integrating alignment methods and alignment information with T -Coffee • Integrating different pair-wise alignment techniques (NW, SW, . . ) • Combining different multiple alignment methods (consensus multiple alignment) • Combining sequence alignment methods with structural alignment techniques • Plug in user knowledge

Matrix extension T-Coffee Tree-based Consistency Objective Function For alignm. Ent Evaluation Cedric Notredame Des Higgins Jaap Heringa J. Mol. Biol. , 302, 205 -217; 2000
Using different sources of alignment information Clustal Structure alignments Dialign Lalign Manual T-Coffee

Search matrix extension – alignment transitivity
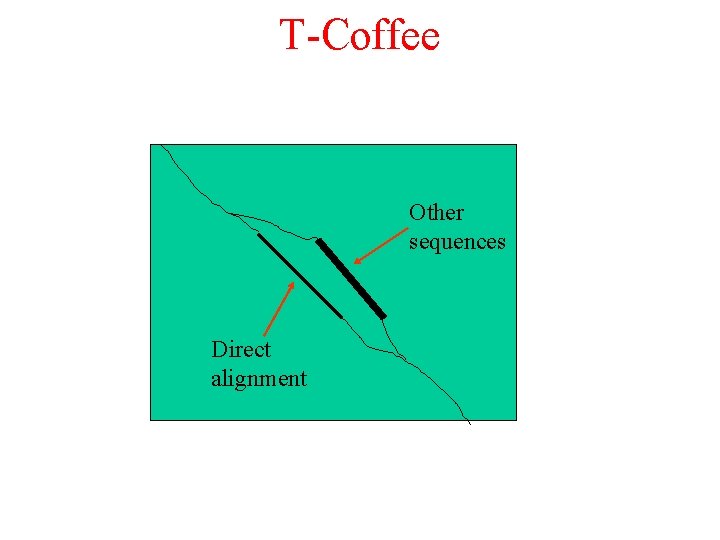
T-Coffee Other sequences Direct alignment
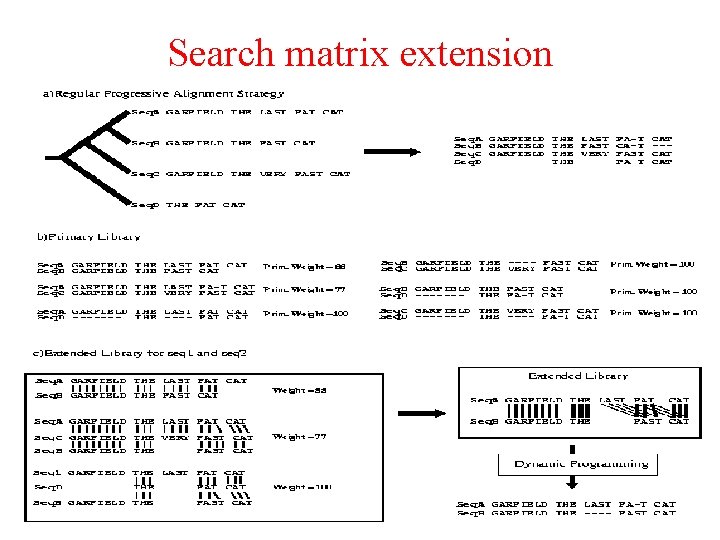
Search matrix extension

but. . . T-COFFEE (V 1. 23) multiple sequence alignment Flavodoxin-che. Y 1 fx 1 FLAV_DESVH FLAV_DESGI FLAV_DESSA FLAV_DESDE 4 fxn FLAV_MEGEL FLAV_CLOAB 2 fcr FLAV_ENTAG FLAV_ANASP FLAV_AZOVI FLAV_ECOLI 3 chy ----PKALIVYGSTTGNTEYTAETIARQLANAG-YEVDSRDAASVE-AGGLFEGFDLVLLGCSTWGDDSIE------LQDDFIPL-FDSLEETGAQGRK-------MPKALIVYGSTTGNTEYTAETIARELADAG-YEVDSRDAASVE-AGGLFEGFDLVLLGCSTWGDDSIE------LQDDFIPL-FDSLEETGAQGRK-------MPKALIVYGSTTGNTEGVAEAIAKTLNSEG-METTVVNVADVT-APGLAEGYDVVLLGCSTWGDDEIE------LQEDFVPL-YEDLDRAGLKDKK-------MSKSLIVYGSTTGNTETAAEYVAEAFENKE-IDVELKNVTDVS-VADLGNGYDIVLFGCSTWGEEEIE------LQDDFIPL-YDSLENADLKGKK-------MSKVLIVFGSSTGNTESIAQKLEELIAAGG-HEVTLLNAADAS-AENLADGYDAVLFGCSAWGMEDLE------MQDDFLSL-FEEFNRFGLAGRK-----MKIVYWSGTGNTEKMAELIAKGIIESG-KDVNTINVSDVN-IDELL-NEDILILGCSAMGDEVLE-------ESEFEPF-IEEIS-TKISGKK-----MVEIVYWSGTGNTEAMANEIEAAVKAAG-ADVESVRFEDTN-VDDVA-SKDVILLGCPAMGSEELE-------DSVVEPF-FTDLA-PKLKGKK----MKISILYSSKTGKTERVAKLIEEGVKRSGNIEVKTMNLDAVD-KKFLQ-ESEGIIFGTPTYYAN-----ISWEMKKW-IDESSEFNLEGKL-----KIGIFFSTSTGNTTEVADFIGKTLGAKA---DAPIDVDDVTDPQAL-KDYDLLFLGAPTWNTGA----DTERSGTSWDEFLYDKLPEVDMKDLP-------MATIGIFFGSDTGQTRKVAKLIHQKLDGIA---DAPLDVRRAT-REQF-LSYPVLLLGTPTLGDGELPGVEAGSQYDSWQEF-TNTLSEADLTGKT-------SKKIGLFYGTQTGKTESVAEIIRDEFGNDV---VTLHDVSQAE-VTDL-NDYQYLIIGCPTWNIGEL----QSDWEGL-YSELDDVDFNGKL----AKIGLFFGSNTGKTRKVAKSIKKRFDDET-M-SDALNVNRVS-AEDF-AQYQFLILGTPTLGEGELPGLSSDCENESWEEF-LPKIEGLDFSGKT----AITGIFFGSDTGNTENIAKMIQKQLGKDV---ADVHDIAKSS-KEDL-EAYDILLLGIPTWYYGEA----QCDWDDF-FPTLEEIDFNGKL----ADKELKFLVVD--DFSTMRRIVRNLLKELGFN-NVE-EAEDGVDALNKLQ-AGGYGFVISDWNMPNMDGLE-------LLKTIRADGAMSALPVLMV : . . . : : ---------VACFGCGDSS--YEYFCGA-VDAIEEKLKNLGAEIVQDG---------------------LRIDGDPRAA--RDDIVGWAHDVRGAI------------VGVFGCGDSS--YTYFCGA-VDVIEKKAEELGATLVASS-----------LKIDGEPDSA----EVLDWAREVLARV--------VSVFGCGDSD--YTYFCGA-VDAIEEKLEKMGAVVIGDS-----------LKIDGDPE----RDEIVSWGSGIADKI--------VAAFASGDQE--YEHFCGA-VPAIEERAKELGATIIAEG-----------LKMEGDASND--PEAVASFAEDVLKQL--------VALFGS------YGWGDGKWMRDFEERMNGYGCVVVETP-----------LIVQNEPD--EAEQDCIEFGKKIANI---------VGLFGS------YGWGSGEWMDAWKQRTEDTGATVIGTA-----------IV--NEMP--DNAPECKELGEAAAKA---------GAAFSTANSI--AGGSDIA-LLTILNHLMVKGMLVY----SGGVAFGKPKTHLGYVHINEIQENEDENARIFGERIANKVKQIF----------VAIFGLGDAEGYPDNFCDA-IEEIHDCFAKQGAKPVGFSNPDDYDYEESKSVRDG-KFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-------VALFGLGDQLNYSKNFVSA-MRILYDLVIARGACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSWLEKLKPAVL--------VAYFGTGDQIGYADNFQDA-IGILEEKISQRGGKTVGYWSTDGYDFNDSKALRNG-KFVGLALDEDNQSDLTDDRIKSWVAQLKSEFGL-------VALFGLGDQVGYPENYLDA-LGELYSFFKDRGAKIVGSWSTDGYEFESSEAVVDG-KFVGLALDLDNQSGKTDERVAAWLAQIAPEFGLSL------VALFGCGDQEDYAEYFCDA-LGTIRDIIEPRGATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKWVKQISEELHLDEILNA TAEAKKENIIAAAQAGASGYVVKPFT---AATLEEKLNKIFEKLGM-----------------------------.
Multiple alignment methods q Multi-dimensional dynamic programming > extension of pairwise sequence alignment. q Progressive alignment > incorporates phylogenetic information to guide the alignment process q Iterative alignment > correct for problems with progressive alignment by repeatedly realigning subgroups of sequence
- Slides: 35