Introduction to bioinformatics Lecture 7 Multiple sequence alignment
Introduction to bioinformatics Lecture 7 Multiple sequence alignment (1)
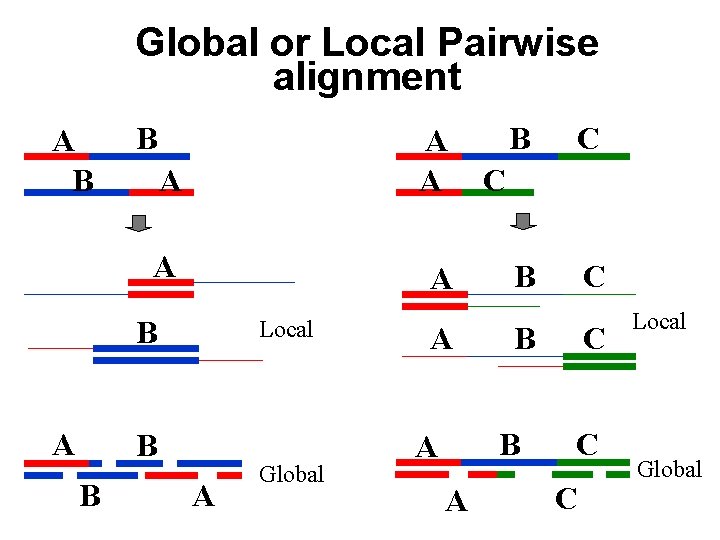
Global or Local Pairwise alignment A B B A A A B A Local B B A Global A B C C B B A A C C C Local Global
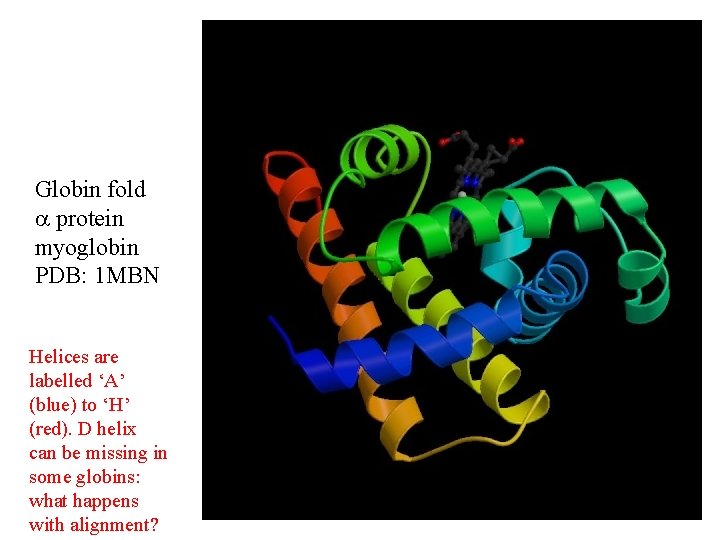
Globin fold protein myoglobin PDB: 1 MBN Helices are labelled ‘A’ (blue) to ‘H’ (red). D helix can be missing in some globins: what happens with alignment?
sandwich protein immunoglobulin PDB: 7 FAB
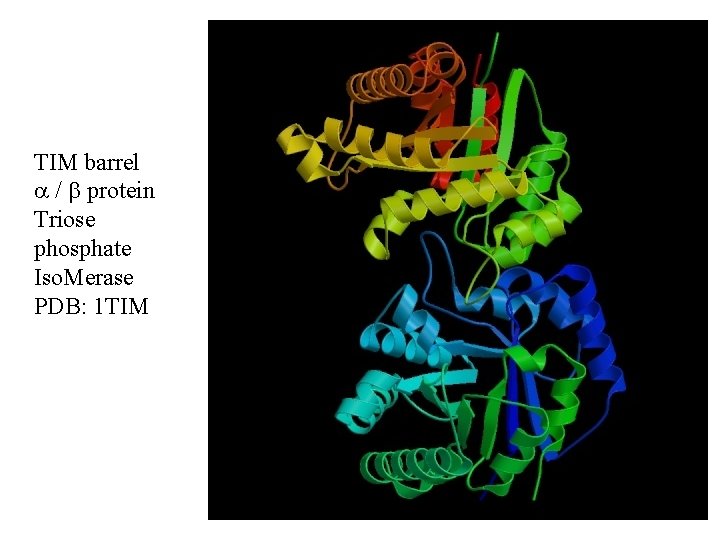
TIM barrel / protein Triose phosphate Iso. Merase PDB: 1 TIM
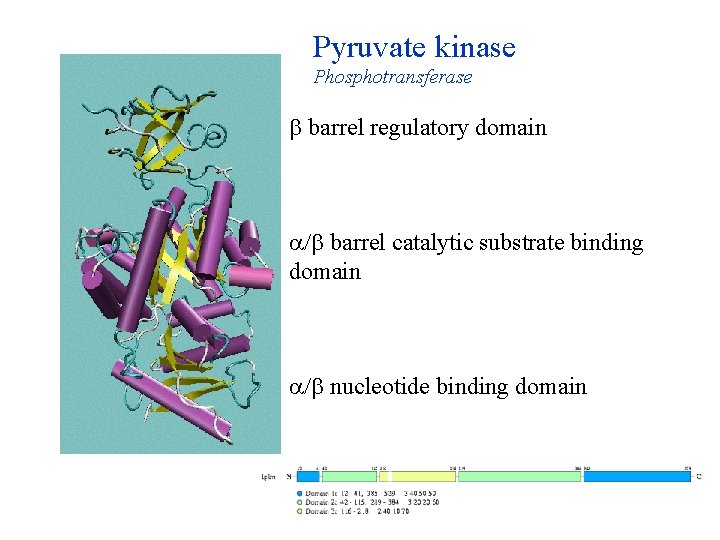
Pyruvate kinase Phosphotransferase barrel regulatory domain / barrel catalytic substrate binding domain / nucleotide binding domain

What does this mean for alignments? n n Alignments need to be able to skip secondary structural elements to complete domains (i. e. putting gaps opposite these motifs in the shorter sequence). Depending on gap penalties chosen, the algorithm might have difficulty with making such long gaps (for example when using high affine gap penalties), resulting in incorrect alignment.

There are three kinds of pairwise alignments n n n Global alignment – align all residues in both sequences; all gaps are penalised Semi-global alignment – align all residues in both sequences; end gaps are not penalised (zero end gap penalties) Local alignment – align part of each sequence; end gaps are not applicable
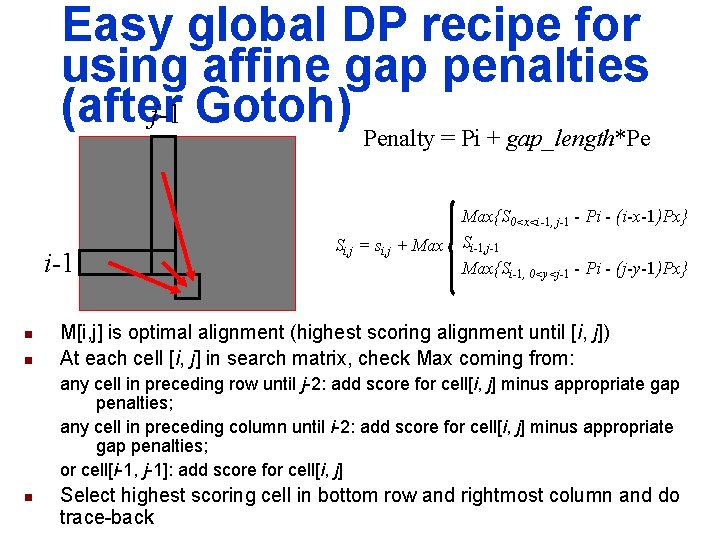
Easy global DP recipe for using affine gap penalties (after j-1 Gotoh) Penalty = Pi + gap_length*Pe i-1 n n Max{S 0<x<i-1, j-1 - Pi - (i-x-1)Px} Si, j = si, j + Max Si-1, j-1 Max{Si-1, 0<y<j-1 - Pi - (j-y-1)Px} M[i, j] is optimal alignment (highest scoring alignment until [i, j]) At each cell [i, j] in search matrix, check Max coming from: any cell in preceding row until j-2: add score for cell[i, j] minus appropriate gap penalties; any cell in preceding column until i-2: add score for cell[i, j] minus appropriate gap penalties; or cell[i-1, j-1]: add score for cell[i, j] n Select highest scoring cell in bottom row and rightmost column and do trace-back
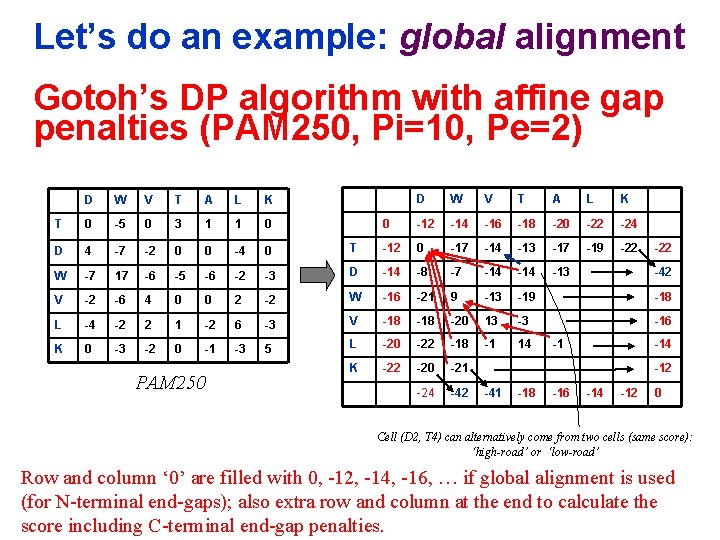
Let’s do an example: global alignment Gotoh’s DP algorithm with affine gap penalties (PAM 250, Pi=10, Pe=2) D W V T A L K 0 -12 -14 -16 -18 -20 -22 -24 T -12 0 -17 -14 -13 -17 -19 -22 -3 D -14 -8 -7 -14 -13 2 -2 W -16 -21 9 -13 -19 -18 -2 6 -3 V -18 -20 13 -3 -16 -1 -3 5 L -20 -22 -18 -1 14 K -22 -20 -21 -24 -42 D W V T A L K T 0 -5 0 3 1 1 0 D 4 -7 -2 0 0 -4 0 W -7 17 -6 -5 -6 -2 V -2 -6 4 0 0 L -4 -2 2 1 K 0 -3 -2 0 PAM 250 -22 -42 -1 -14 -12 -41 -18 -16 -14 -12 0 Cell (D 2, T 4) can alternatively come from two cells (same score): ‘high-road’ or ‘low-road’ Row and column ‘ 0’ are filled with 0, -12, -14, -16, … if global alignment is used (for N-terminal end-gaps); also extra row and column at the end to calculate the score including C-terminal end-gap penalties.
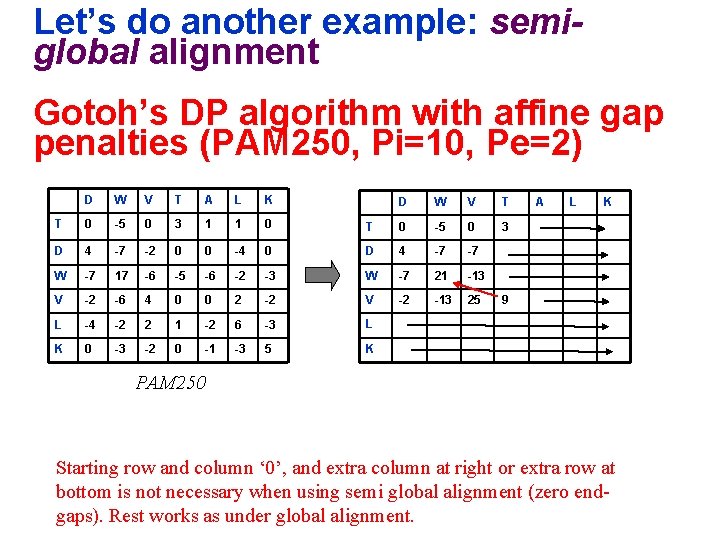
Let’s do another example: semiglobal alignment Gotoh’s DP algorithm with affine gap penalties (PAM 250, Pi=10, Pe=2) D W V T A L K T 0 -5 0 3 1 1 0 D 4 -7 -2 0 0 -4 W -7 17 -6 -5 -6 V -2 -6 4 0 L -4 -2 2 K 0 -3 -2 D W V T T 0 -5 0 3 0 D 4 -7 -7 -2 -3 W -7 21 -13 0 2 -2 V -2 -13 25 1 -2 6 -3 L 0 -1 -3 5 K A L K 9 PAM 250 Starting row and column ‘ 0’, and extra column at right or extra row at bottom is not necessary when using semi global alignment (zero endgaps). Rest works as under global alignment.
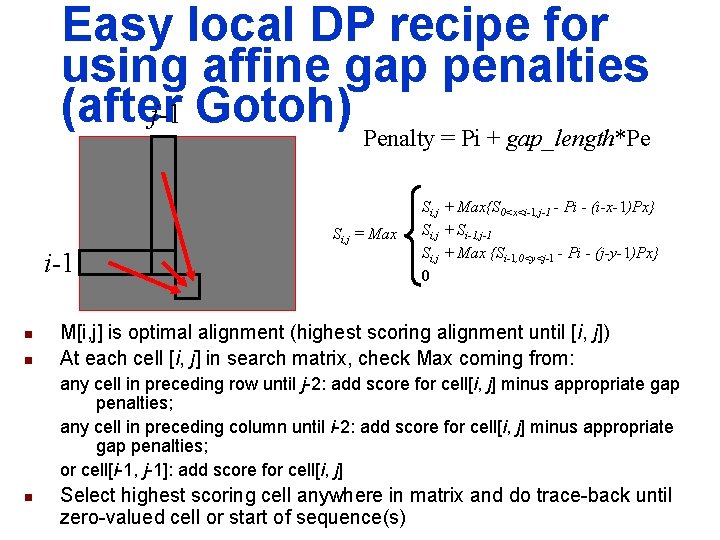
Easy local DP recipe for using affine gap penalties (after j-1 Gotoh) Penalty = Pi + gap_length*Pe Si, j = Max i-1 n n Si, j + Max{S 0<x<i-1, j-1 - Pi - (i-x-1)Px} Si, j + Si-1, j-1 Si, j + Max {Si-1, 0<y<j-1 - Pi - (j-y-1)Px} 0 M[i, j] is optimal alignment (highest scoring alignment until [i, j]) At each cell [i, j] in search matrix, check Max coming from: any cell in preceding row until j-2: add score for cell[i, j] minus appropriate gap penalties; any cell in preceding column until i-2: add score for cell[i, j] minus appropriate gap penalties; or cell[i-1, j-1]: add score for cell[i, j] n Select highest scoring cell anywhere in matrix and do trace-back until zero-valued cell or start of sequence(s)
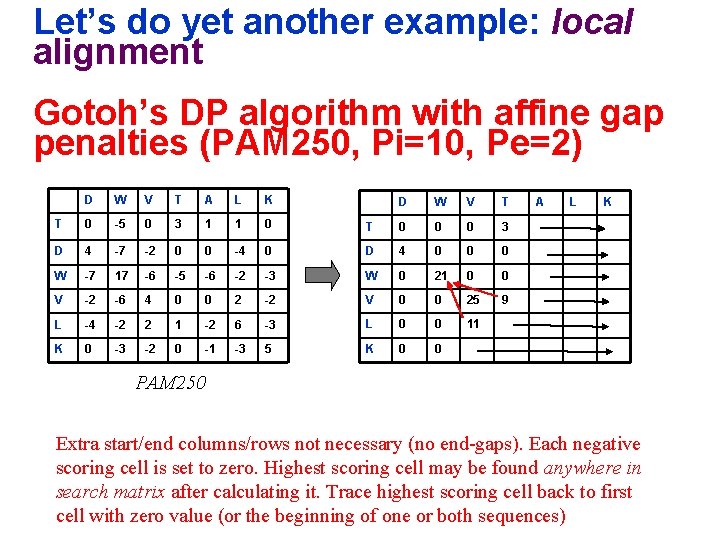
Let’s do yet another example: local alignment Gotoh’s DP algorithm with affine gap penalties (PAM 250, Pi=10, Pe=2) D W V T A L K T 0 -5 0 3 1 1 0 D 4 -7 -2 0 0 -4 W -7 17 -6 -5 -6 V -2 -6 4 0 L -4 -2 2 K 0 -3 -2 D W V T T 0 0 0 3 0 D 4 0 0 0 -2 -3 W 0 21 0 0 0 2 -2 V 0 0 25 9 1 -2 6 -3 L 0 0 11 0 -1 -3 5 K 0 0 A L K PAM 250 Extra start/end columns/rows not necessary (no end-gaps). Each negative scoring cell is set to zero. Highest scoring cell may be found anywhere in search matrix after calculating it. Trace highest scoring cell back to first cell with zero value (or the beginning of one or both sequences)

For your first exam D 1: Make sure you understand can carry out Gotoh’s algorithm for global, semiglobal and local alignment! This is the most general Dynamic Programming (DP) algorithm (and perhaps the easiest to understand) Gotoh, O. An Improved Algorithm for Matching Biological Sequences. J. Mol. Biol. , 162, pp.

Pairwise alignment n n n Now we know how to do it: How do we get a multiple alignment (three or more sequences)? Multiple alignment: much greater combinatorial explosion than with pairwise alignment…. .
Multiple sequence alignment (MSA) Why n n n One of he most important means to find out about: n Conservation patterns leading to functional clues n Possible protein structure Multiple sequence alignment contains far more information about conservation than pairwise alignment Many bioinformatics methods use MSA as input: e. g. secondary structure prediction (later lecture)

Multiple sequence alignment Wanted n n n Quality Programs need to be fully automatic for genomic pipelines With available genomes (data explosion), speed becomes crucial
Simultaneous Multiple alignment Multi-dimensional dynamic programming (Murata et al. 1985)
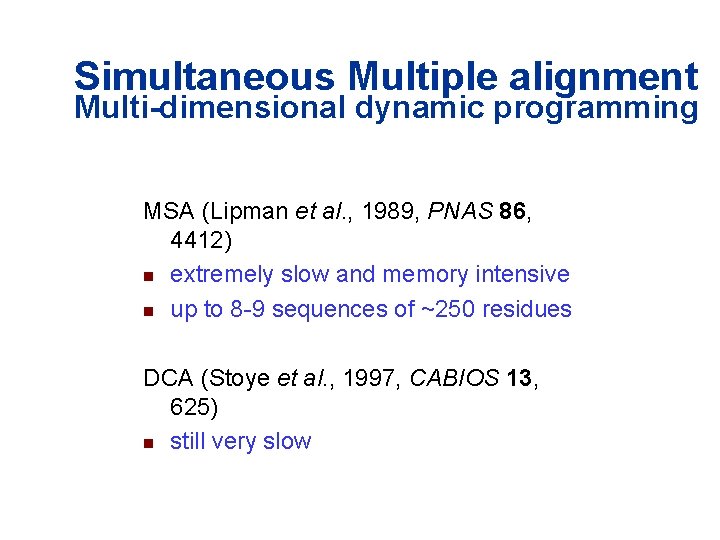
Simultaneous Multiple alignment Multi-dimensional dynamic programming MSA (Lipman et al. , 1989, PNAS 86, 4412) n extremely slow and memory intensive n up to 8 -9 sequences of ~250 residues DCA (Stoye et al. , 1997, CABIOS 13, 625) n still very slow
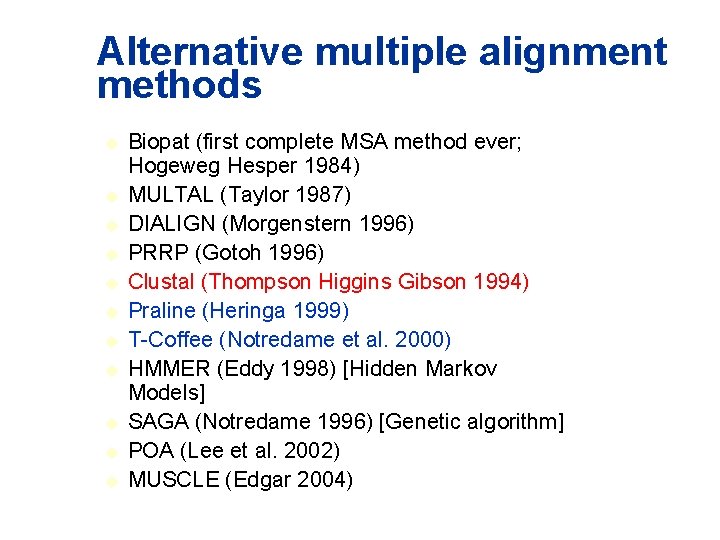
Alternative multiple alignment methods u u u Biopat (first complete MSA method ever; Hogeweg Hesper 1984) MULTAL (Taylor 1987) DIALIGN (Morgenstern 1996) PRRP (Gotoh 1996) Clustal (Thompson Higgins Gibson 1994) Praline (Heringa 1999) T-Coffee (Notredame et al. 2000) HMMER (Eddy 1998) [Hidden Markov Models] SAGA (Notredame 1996) [Genetic algorithm] POA (Lee et al. 2002) MUSCLE (Edgar 2004)

The following three slides are examples of multiple alignments of 13 flavodoxin and 1 che. Y sequence (PDB code 3 chy). The che. Y sequence is a very distant relative of the flavodoxin family, but has the same basic fold

CLUSTAL X (1. 64 b) multiple sequence alignment Flavodoxin-che. Y 1 fx 1 FLAV_DESVH FLAV_DESGI FLAV_DESSA FLAV_DESDE FLAV_CLOAB FLAV_MEGEL 4 fxn FLAV_ANASP FLAV_AZOVI 2 fcr FLAV_ENTAG FLAV_ECOLI 3 chy -PKALIVYGSTTGNTEYTAETIARQLANAG-Y-EVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSIE------LQDDFIPLFD-SLEETGAQGRK MPKALIVYGSTTGNTEYTAETIARELADAG-Y-EVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSIE------LQDDFIPLFD-SLEETGAQGRK MPKALIVYGSTTGNTEGVAEAIAKTLNSEG-M-ETTVVNVADVTAPGLAEGYDVVLLGCSTWGDDEIE------LQEDFVPLYE-DLDRAGLKDKK MSKSLIVYGSTTGNTETAAEYVAEAFENKE-I-DVELKNVTDVSVADLGNGYDIVLFGCSTWGEEEIE------LQDDFIPLYD-SLENADLKGKK MSKVLIVFGSSTGNTESIAQKLEELIAAGG-H-EVTLLNAADASAENLADGYDAVLFGCSAWGMEDLE------MQDDFLSLFE-EFNRFGLAGRK -MKISILYSSKTGKTERVAKLIEEGVKRSGNI-EVKTMNLDAVDKKFLQE-SEGIIFGTPTYYAN-----ISWEMKKWID-ESSEFNLEGKL --MVEIVYWSGTGNTEAMANEIEAAVKAAG-A-DVESVRFEDTNVDDVAS-KDVILLGCPAMGSE--E------LEDSVVEPFF-TDLAPKLKGKK ---MKIVYWSGTGNTEKMAELIAKGIIESG-K-DVNTINVSDVNIDELLN-EDILILGCSAMGDE--V------LEESEFEPFI-EEISTKISGKK SKKIGLFYGTQTGKTESVAEIIRDEFGNDVVT----LHDVSQAEVTDLND-YQYLIIGCPTWNIGELQ---SD-----WEGLYS-ELDDVDFNGKL -AKIGLFFGSNTGKTRKVAKSIKKRFDDETMSD---ALNVNRVSAEDFAQ-YQFLILGTPTLGEGELPGLSSDCENESWEEFLP-KIEGLDFSGKT --KIGIFFSTSTGNTTEVADFIGKTLGAKADAP---IDVDDVTDPQALKD-YDLLFLGAPTWNTGADTERSGT----SWDEFLYDKLPEVDMKDLP MATIGIFFGSDTGQTRKVAKLIHQKLDGIADAP---LDVRRATREQFLS--YPVLLLGTPTLGDGELPGVEAGSQYDSWQEFTN-TLSEADLTGKT -AITGIFFGSDTGNTENIAKMIQKQLGKDVAD----VHDIAKSSKEDLEA-YDILLLGIPTWYYGEAQ-CD-------WDDFFP-TLEEIDFNGKL --ADKELKFLVVDDFSTMRRIVRNLLKELG----FNNVEEAEDGVDALN------KLQAGGYGFV--I------SDWNMPNMDG-LELLKTIR--. . : 1 fx 1 FLAV_DESVH FLAV_DESGI FLAV_DESSA FLAV_DESDE FLAV_CLOAB FLAV_MEGEL 4 fxn FLAV_ANASP FLAV_AZOVI 2 fcr FLAV_ENTAG FLAV_ECOLI 3 chy VACFGCGDSSYEYF--CGAVDAIEEKLKNLGAEIVQDG----------------LRIDGDPRAARDDIVGWAHDVRGAI--------------VGVFGCGDSSYTYF--CGAVDVIEKKAEELGATLVASS--------LKIDGEPDSAE--VLDWAREVLARV-------VSVFGCGDSDYTYF--CGAVDAIEEKLEKMGAVVIGDS--------LKIDGDPERDE--IVSWGSGIADKI-------VAAFASGDQEYEHF--CGAVPAIEERAKELGATIIAEG--------LKMEGDASNDPEAVASFAEDVLKQL-------GAAFSTANSIAGGS--DIALLTILNHLMVKGMLVYSGGVA----FGKPKTHLGYVHINEIQENEDENARIFGERIANKVKQIF-----VGLFGSYGWGSGE-----WMDAWKQRTEDTGATVIGTA--------IVN-EMPDNAPECKE-LGEAAAKA--------VALFGSYGWGDGK-----WMRDFEERMNGYGCVVVETP--------LIVQNEPDEAEQDCIEFGKKIANI--------VAYFGTGDQIGYADNFQDAIGILEEKISQRGGKTVGYWSTDGYDFNDSKALR-NGKFVGLALDEDNQSDLTDDRIKSWVAQLKSEFGL-----VALFGLGDQVGYPENYLDALGELYSFFKDRGAKIVGSWSTDGYEFESSEAVV-DGKFVGLALDLDNQSGKTDERVAAWLAQIAPEFGLSL---VAIFGLGDAEGYPDNFCDAIEEIHDCFAKQGAKPVGFSNPDDYDYEESKSVR-DGKFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-----VALFGLGDQLNYSKNFVSAMRILYDLVIARGACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSWLEKLKPAVL------VALFGCGDQEDYAEYFCDALGTIRDIIEPRGATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKWVKQISEELHLDEILNA AD--GAMSALPVL-----MVTAEAKKENIIAAAQAGAS--------GYV-VKPFTAATLEEKLNKIFEKLGM-------. . : . .

Flavodoxin-che. Y: Global Preprocessing (prepro 1500) 1 fx 1 FLAV_DESDE FLAV_DESVH FLAV_DESSA FLAV_DESGI 2 fcr FLAV_AZOVI FLAV_ENTAG FLAV_ANASP FLAV_ECOLI 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy -PKALIVYGSTTGNT-EYTAETIARQLANAG-YEVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSI------ELQDDFIPLF-DSLEETGAQGRKVACF MSKVLIVFGSSTGNT-ESIa. QKLEELIAAGG-HEVTLLNAADASAENLADGYDAVLFg. CSAWGMEDL------EMQDDFLSLF-EEFNRFGLAGRKVAAf MPKALIVYGSTTGNT-EYTa. ETIARELADAG-YEVDSRDAASVEAGGLFEGFDLVLLg. CSTWGDDSI------ELQDDFIPLF-DSLEETGAQGRKVACf MSKSLIVYGSTTGNT-ETAa. EYVAEAFENKE-IDVELKNVTDVSVADLGNGYDIVLFg. CSTWGEEEI------ELQDDFIPLY-DSLENADLKGKKVSVf MPKALIVYGSTTGNT-EGVa. EAIAKTLNSEG-METTVVNVADVTAPGLAEGYDVVLLg. CSTWGDDEI------ELQEDFVPLY-EDLDRAGLKDKKVGVf --KIGIFFSTSTGNT-TEVADFIGKTLGA---KADAPIDVDDVTDPQALKDYDLLFLGAPTWNTG----ADTERSGTSWDEFLYDKLPEVDMKDLPVAIF -AKIGLFFGSNTGKT-RKVa. KSIKKRFDDET-MSDA-LNVNRVS-AEDFAQYQFLILg. TPTLGEGELPGLSSDCENESWEEFL-PKIEGLDFSGKTVALf MATIGIFFGSDTGQT-RKVa. KLIHQKLDG---IADAPLDVRRAT-REQFLSYPVLLLg. TPTLGDGELPGVEAGSQYDSWQEFT-NTLSEADLTGKTVALf SKKIGLFYGTQTGKT-ESVa. EIIRDEFGN---DVVTLHDVSQAE-VTDLNDYQYLIIg. CPTWNIGEL----QSDWEGLY-SELDDVDFNGKLVAYf -AITGIFFGSDTGNT-ENIa. KMIQKQLGK---DVADVHDIAKSS-KEDLEAYDILLLg. IPTWYYGE----AQCDWDDFF-PTLEEIDFNGKLVALf -MK--IVYWSGTGNT-EKMAELIAKGIIESG-KDVNTINVSDVNIDELL-NEDILILGCSAMGDEVL-------EESEFEPFI-EEIS-TKISGKKVALF MVE--IVYWSGTGNT-EAMa. NEIEAAVKAAG-ADVESVRFEDTNVDDVA-SKDVILLg. CPAMGSEEL-------EDSVVEPFF-TDLA-PKLKGKKVGLf -MKISILYSSKTGKT-ERVa. KLIEEGVKRSGNIEVKTMNLDAVD-KKFLQESEGIIFg. TPTYYAN-----ISWEMKKWI-DESSEFNLEGKLGAAf ADKELKFLVVDDFSTMRRIVRNLLKELGFN--NVEEAEDGVDALNKLQAGGYGFVI---SDWNMPNM-----DGLELL-KTIRADGAMSALPVLM 1 fx 1 FLAV_DESDE FLAV_DESVH FLAV_DESSA FLAV_DESGI 2 fcr FLAV_AZOVI FLAV_ENTAG FLAV_ANASP FLAV_ECOLI 4 fxn FLAV_MEGEL FLAV_CLOAB 3 chy GCGDS-SY-EYFCGA-VDAIEEKLKNLGAEIVQD-----------GLRIDGD--PRAARDDIVGWAHDVRGAI-------ASGDQ-EY-EHFCGA-VPAIEERAKELg. ATIIAE-----------GLKMEGD--ASNDPEAVASf. AEDVLKQL-------GCGDS-SY-EYFCGA-VDAIEEKLKNLg. AEIVQD-----------GLRIDGD--PRAARDDIVGw. AHDVRGAI-------GCGDS-DY-TYFCGA-VDAIEEKLEKMg. AVVIGD-----------SLKIDGD--PE--RDEIVSw. GSGIADKI-------GCGDS-SY-TYFCGA-VDVIEKKAEELg. ATLVAS-----------SLKIDGE--PD--SAEVLDw. AREVLARV-------GLGDAEGYPDNFCDA-IEEIHDCFAKQGAKPVGFSNPDDYDYEESKS-VRDGKFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-----GLGDQVGYPENYLDA-LGELYSFFKDRg. AKIVGSWSTDGYEFESSEA-VVDGKFVGLALDLDNQSGKTDERVAAw. LAQIAPEFGLS--L-GLGDQLNYSKNFVSA-MRILYDLVIARg. ACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSw. LEKLKPAV-L-----GTGDQIGYADNFQDA-IGILEEKISQRg. GKTVGYWSTDGYDFNDSKA-LRNGKFVGLALDEDNQSDLTDDRIKSw. VAQLKSEFGL-----GCGDQEDYAEYFCDA-LGTIRDIIEPRg. ATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKw. VKQISEELHLDEILNA G-----SY-GWGDGKWMRDFEERMNGYGCVVVET-----------PLIVQNE--PDEAEQDCIEFGKKIANI----G-----SY-GWGSGEWMDAWKQRTEDTg. ATVIGT-----------AIVNEM--PDNA-PECKEl. GEAAAKA----STANSIAGGSDIA---LLTILNHLMVKg. MLVYSG----GVAFGKPKTHLGYVHINEIQENEDENARIf. GERi. ANk. VKQIF-----VTAEAKK--ENIIAA-----AQAGAS-------------GYVV-----KPFTAATLEEKLNKIFEKLGM------ Iteration 0 T G SP= 136944. 00 Av. SP= 10. 675 SId= 4009 Av. SId= 0. 313

Flavodoxin-che. Y: Local Pre-processing (locprepro 300) 1 fx 1 FLAV_DESVH FLAV_DESSA FLAV_DESGI FLAV_DESDE 4 fxn FLAV_MEGEL 2 fcr FLAV_ANASP FLAV_AZOVI FLAV_ENTAG FLAV_ECOLI FLAV_CLOAB 3 chy --PKALIVYGSTTGNTEYTAETIARQLANAGYEVDSRDAASVEAGGLFEGFDLVLLGCSTWGDDSI------ELQDDFIPL--FDSLEETGAQGRKVACF -MPKALIVYGSTTGNTEYTa. ETIARELADAGYEVDSRDAASVEAGGLFEGFDLVLLg. CSTWGDDSI------ELQDDFIPL--FDSLEETGAQGRKVACf -MSKSLIVYGSTTGNTETAa. EYVAEAFENKEIDVELKNVTDVSVADLGNGYDIVLFg. CSTWGEEEI------ELQDDFIPL--YDSLENADLKGKKVSVf -MPKALIVYGSTTGNTEGVa. EAIAKTLNSEGMETTVVNVADVTAPGLAEGYDVVLLg. CSTWGDDEI------ELQEDFVPL--YEDLDRAGLKDKKVGVf -MSKVLIVFGSSTGNTESIa. QKLEELIAAGGHEVTLLNAADASAENLADGYDAVLFg. CSAWGMEDL------EMQDDFLSL--FEEFNRFGLAGRKVAAf --MK--IVYWSGTGNTEKMAELIAKGIIESGKDVNTINVSDVNIDELLN-EDILILGCSAMGDEVL------E-ESEFEPF--IEEIS-TKISGKKVALF -MVE--IVYWSGTGNTEAMa. NEIEAAVKAAGADVESVRFEDTNVDDVAS-KDVILLg. CPAMGSEEL------E-DSVVEPF--FTDLA-PKLKGKKVGLf ---KIGIFFSTSTGNTTEVADFIGKTLGAKADAPI--DVDDVTDPQALKDYDLLFLGAPTWNTGAD----TERSGTSWDEFL-YDKLPEVDMKDLPVAIF -SKKIGLFYGTQTGKTESVa. EIIRDEFGNDVVTLH--DVSQAEV-TDLNDYQYLIIg. CPTWNIGEL----QSDWEGL--YSELDDVDFNGKLVAYf --AKIGLFFGSNTGKTRKVa. KSIKKRFDDETMSDA-LNVNRVSA-EDFAQYQFLILg. TPTLGEGELPGLSSDCENESWEEF--LPKIEGLDFSGKTVALf -MATIGIFFGSDTGQTRKVa. KLIHQKLDG--IADAPLDVRRATR-EQFLSYPVLLLg. TPTLGDGELPGVEAGSQYDSWQEF--TNTLSEADLTGKTVALf --AITGIFFGSDTGNTENIa. KMIQKQLGKDVADVH--DIAKSSK-EDLEAYDILLLg. IPTWYYGEA----QCDWDDF--FPTLEEIDFNGKLVALf --MKISILYSSKTGKTERVa. KLIEEGVKRSGNIEVKTMNLDAVDKKFLQESEGIIFg. TPTYYA------NISWEMKKWIDESSEFNLEGKLGAAf ADKELKFLVVDDFSTMRRIVRNLLKELGFNNVEEAEDGVDALNKLQ-AGGYGFVI---SDWNMPNM-----DGLEL--LKTIRADGAMSALPVLM 1 fx 1 FLAV_DESVH FLAV_DESSA FLAV_DESGI FLAV_DESDE 4 fxn FLAV_MEGEL 2 fcr FLAV_ANASP FLAV_AZOVI FLAV_ENTAG FLAV_ECOLI FLAV_CLOAB 3 chy GCGDS--SY-EYFCGA-VD--AIEEKLKNLGAEIVQD-----------GLRID--GDPRAARDDIVGWAHDVRGAI-------GCGDS--SY-EYFCGA-VD--AIEEKLKNLg. AEIVQD-----------GLRID--GDPRAARDDIVGw. AHDVRGAI-------GCGDS--DY-TYFCGA-VD--AIEEKLEKMg. AVVIGD-----------SLKID--GDPE--RDEIVSw. GSGIADKI-------GCGDS--SY-TYFCGA-VD--VIEKKAEELg. ATLVAS-----------SLKID--GEPD--SAEVLDw. AREVLARV-------ASGDQ--EY-EHFCGA-VP--AIEERAKELg. ATIIAE-----------GLKME--GDASNDPEAVASf. AEDVLKQL-------GS------Y-GWGDGKWMR--DFEERMNGYGCVVVET-----------PLIVQ--NEPDEAEQDCIEFGKKIANI----GS------Y-GWGSGEWMD--AWKQRTEDTg. ATVIGT-----------AI-VN--EMPDNA-PECKEl. GEAAAKA----GLGDAE-GYPDNFCDA-IE--EIHDCFAKQGAKPVGFSNPDDYDYEESKSVRD-GKFLGLPLDMVNDQIPMEKRVAGWVEAVVSETGV-----GTGDQI-GYADNFQDA-IG--ILEEKISQRg. GKTVGYWSTDGYDFNDSKALRN-GKFVGLALDEDNQSDLTDDRIKSw. VAQLKSEFGL-----GLGDQV-GYPENYLDA-LG--ELYSFFKDRg. AKIVGSWSTDGYEFESSEAVVD-GKFVGLALDLDNQSGKTDERVAAw. LAQIAPEFGLS--L-GLGDQL-NYSKNFVSA-MR--ILYDLVIARg. ACVVGNWPREGYKFSFSAALLENNEFVGLPLDQENQYDLTEERIDSw. LEKLKPAV-L-----GCGDQE-DYAEYFCDA-LG--TIRDIIEPRg. ATIVGHWPTAGYHFEASKGLADDDHFVGLAIDEDRQPELTAERVEKw. VKQISEELHLDEILNA STANSIAGGSDIALLTILNHLMVKg. MLVYSGGVAFGKPKTHLGYVH-----INEIQENEDENARIf. GERi. ANk. VKQIF-----VTAEA---KKENIIAA------AQAGAS-------------GYVVK-----PFTAATLEEKLNKIFEKLGM------ G
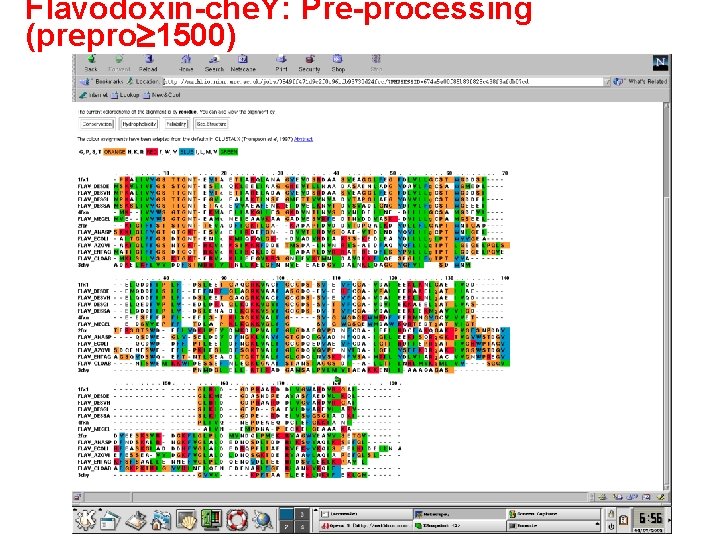
Flavodoxin-che. Y: Pre-processing (prepro 1500)
- Slides: 25