Introduction Medical Application and Recent Advances in Molecular

























- Slides: 67

Introduction, Medical Application and Recent Advances in Molecular Biology and Bioinformatics • Padmini Narayanan, Ph. D. pnarayan@bcm. edu Refresher Course in Molecular Biology and Bioinformatics, 2018

Learning Objectives • Why do you need to attend this series? What can you expect from this course? How can you translate this knowledge in to better patient care? • To provide a solid introduction to molecular biology with an emphasis on techniques and bioinformatic tools that are currently used to investigate the molecular mechanisms and its application to diagnose and treat diseases. • To introduce and give context to all the following lectures of this course

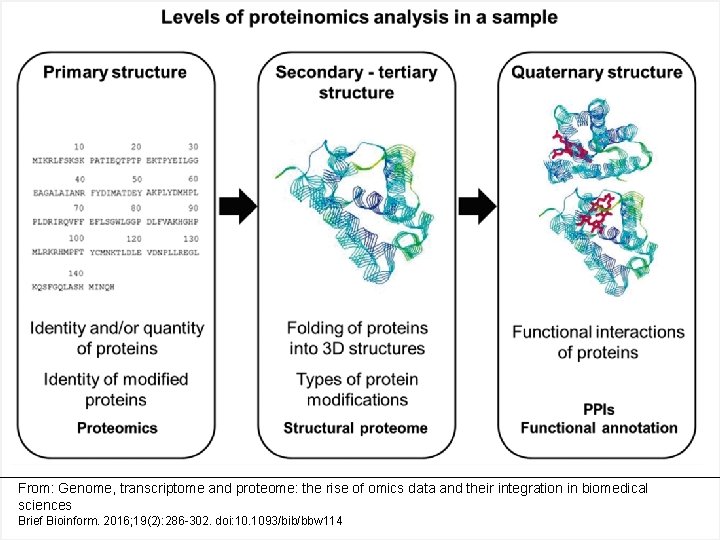
From: Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences Brief Bioinform. 2016; 19(2): 286 -302. doi: 10. 1093/bib/bbw 114

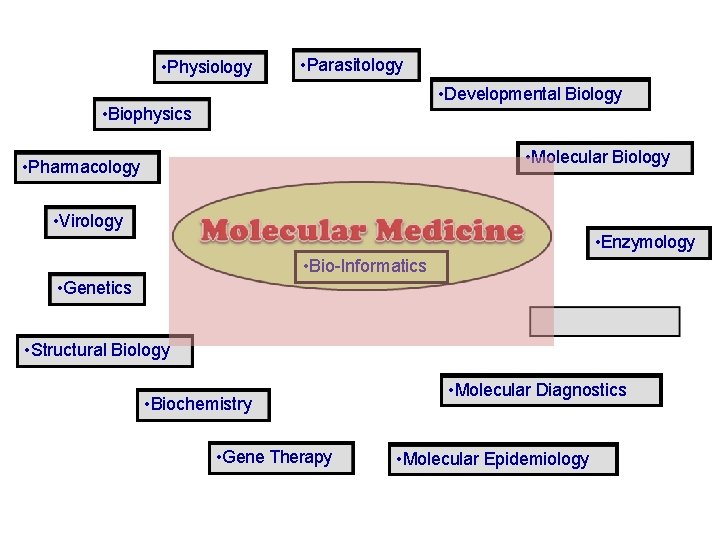
• Physiology • Parasitology • Developmental Biology • Biophysics • Molecular Biology • Pharmacology • Virology • Enzymology • Genetics • Bio-Informatics • Structural Biology • Biochemistry • Gene Therapy • Molecular Diagnostics • Molecular Epidemiology

• Molecular Medicine Molecular Insights · · Identify pathology Find inheritance of disease Find ‘candidate gene’ Screen for mutations Applications • Improved diagnosis of disease • Earlier detection of genetic predisposition to disease (BRCA 1) • Rational drug design • Gene therapy • “targeted/custom drugs" • lindsaydigital. c

• Physiology • Parasitology • Developmental Biology • Biophysics • Molecular Biology • Pharmacology • Virology • Enzymology • Bio-Informatics • Genetics • Structural Biology • Biochemistry • Gene Therapy • Molecular Diagnostics • Molecular Epidemiology

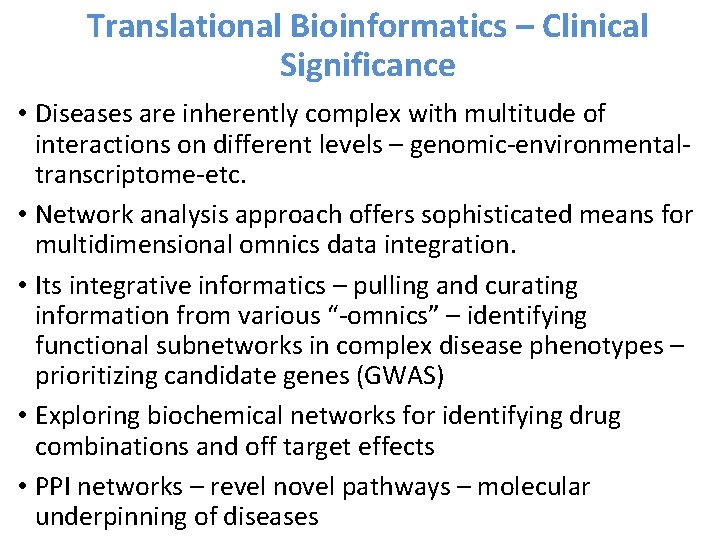
Translational Bioinformatics – Clinical Significance • Diseases are inherently complex with multitude of interactions on different levels – genomic-environmentaltranscriptome-etc. • Network analysis approach offers sophisticated means for multidimensional omnics data integration. • Its integrative informatics – pulling and curating information from various “-omnics” – identifying functional subnetworks in complex disease phenotypes – prioritizing candidate genes (GWAS) • Exploring biochemical networks for identifying drug combinations and off target effects • PPI networks – revel novel pathways – molecular underpinning of diseases

Translational Bioinformatics – Clinical Significance • Using this approach – new methods/ models can be developed • Better understanding of different aspects of the disease progression • Generate new diagnostic tools – biomarkers – prognostic and drug targets • Valid hypothesis generation , targeted therapy – personalized medicine

RESOURCES • The Cancer Genomic Atlas (TCGA) • Therapeutically Applkicable Research to generate Effective Treatments (TARGET) • Cancer Target and Driver Discovery Network (CTD 2) • NCI Genomic Data commons (GDC)

PROCESSES, TOOLS AND APPLICATION

Gene and Gene regulatory units Replication Transcription Splicing Transport, Stability Translation Structure, Transport, Stability Posttranslational modifications Interactions Activity Downstream signaling Structural support https: //www. dnalc. org/resources/animations/

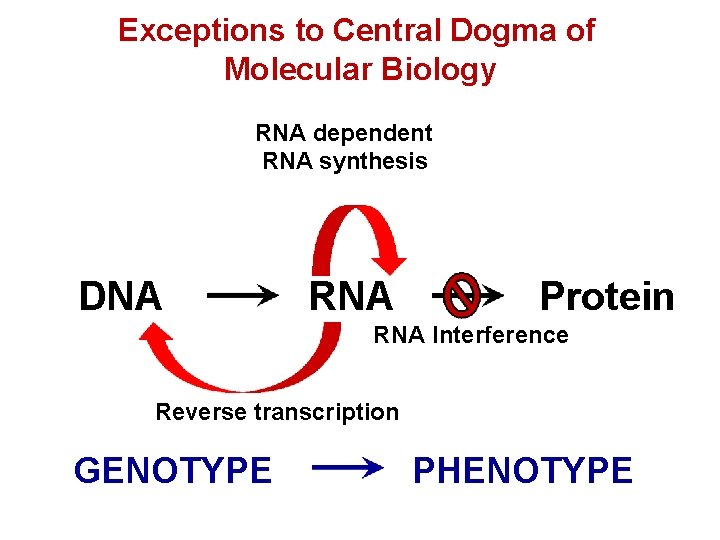
Exceptions to Central Dogma of Molecular Biology RNA dependent RNA synthesis DNA RNA Protein RNA Interference Reverse transcription GENOTYPE PHENOTYPE

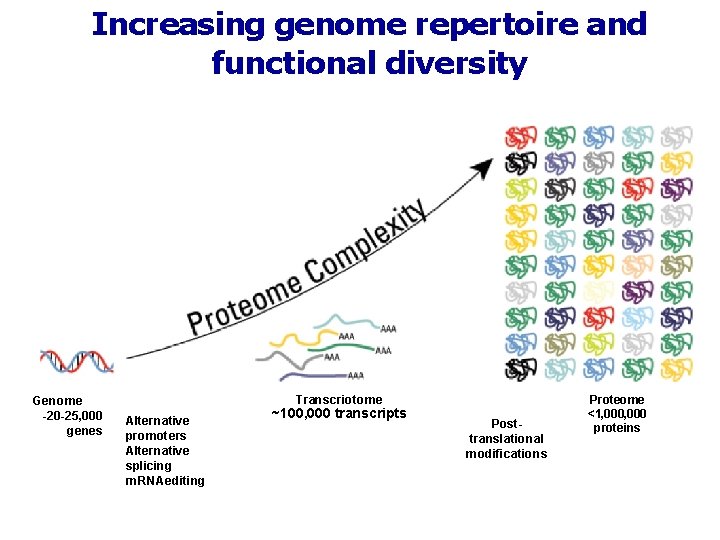
Increasing genome repertoire and functional diversity Genome -20 -25, 000 genes Transcriotome Alternative promoters Alternative splicing rn. RNAediting ~100, 000 transcripts Posttranslational modifications Proteome <1, 000 proteins

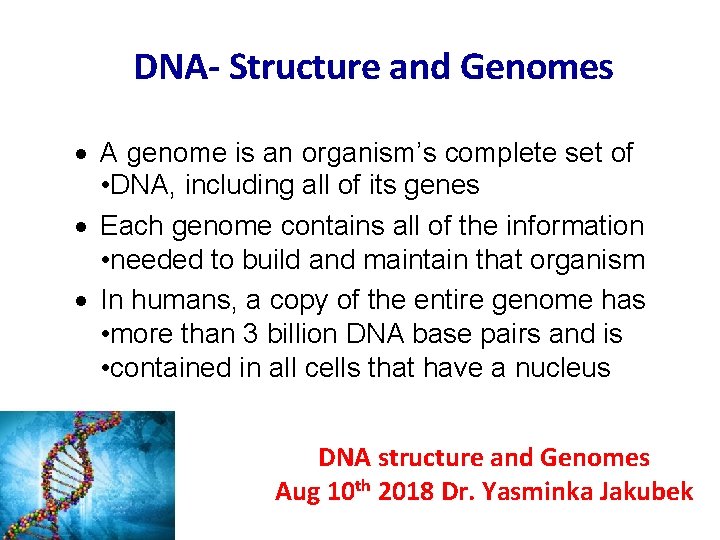
DNA- Structure and Genomes · A genome is an organism’s complete set of • DNA, including all of its genes · Each genome contains all of the information • needed to build and maintain that organism · In humans, a copy of the entire genome has • more than 3 billion DNA base pairs and is • contained in all cells that have a nucleus DNA structure and Genomes Aug 10 th 2018 Dr. Yasminka Jakubek

Human Genome, 3 x 109 bp packaged into chromosomes Ø <3 % is expressed as proteins!! Ø ~80% is transcribed Ø What does the transcripts do? ? Regulatory function Ø remaining 98– 99% (noncoding regions) holds structural and functional relevance http: //www. sciencemag. org/news/2012/09/humangenome-much-more-just-genes

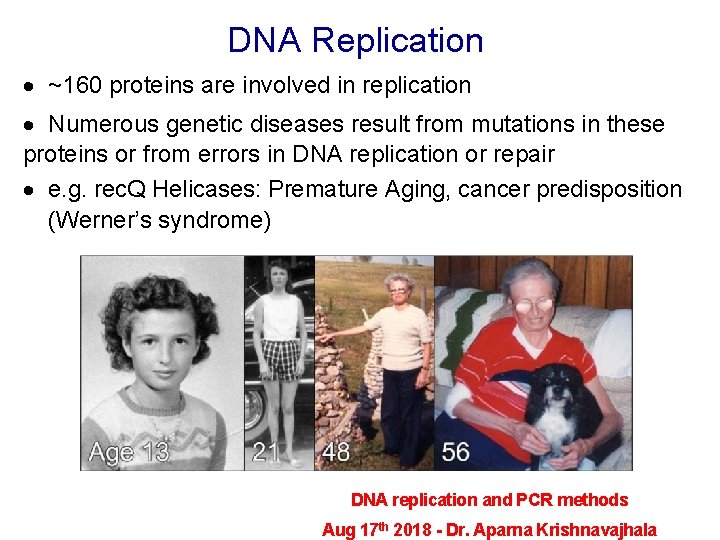
DNA Replication · ~160 proteins are involved in replication · Numerous genetic diseases result from mutations in these proteins or from errors in DNA replication or repair · e. g. rec. Q Helicases: Premature Aging, cancer predisposition (Werner’s syndrome) DNA replication and PCR methods Aug 17 th 2018 - Dr. Aparna Krishnavajhala

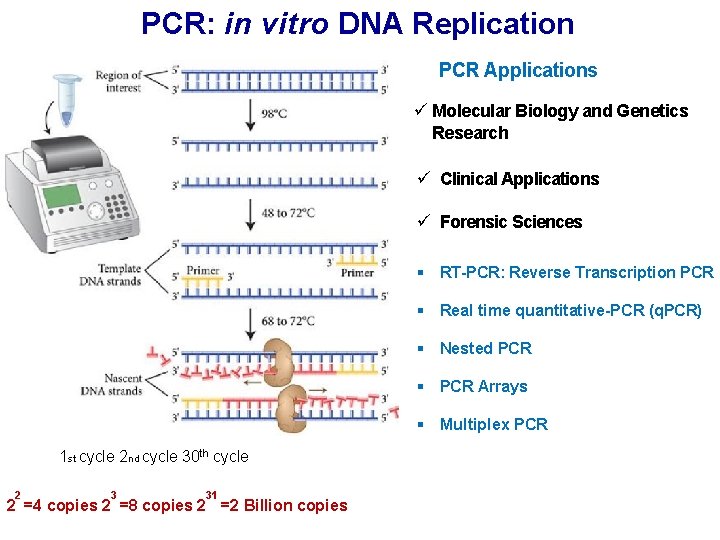
PCR: in vitro DNA Replication PCR Applications ü Molecular Biology and Genetics Research ü Clinical Applications ü Forensic Sciences § RT-PCR: Reverse Transcription PCR § Real time quantitative-PCR (q. PCR) § Nested PCR § PCR Arrays § Multiplex PCR 1 st cycle 2 nd cycle 30 th cycle 2 3 31 2 =4 copies 2 =8 copies 2 =2 Billion copies

DNA Sequencing · · Very important tool for biologists Sequence of genes Positioning of genes Sequences of regulatory regions Applications: üHealth Care üDiagnostics üForensics (DNA fingerprinting) üAgriculture üResearch Next generation DNA sequencing methods Aug 24 th 2018 Dr. Devon Marie Fitzgerald

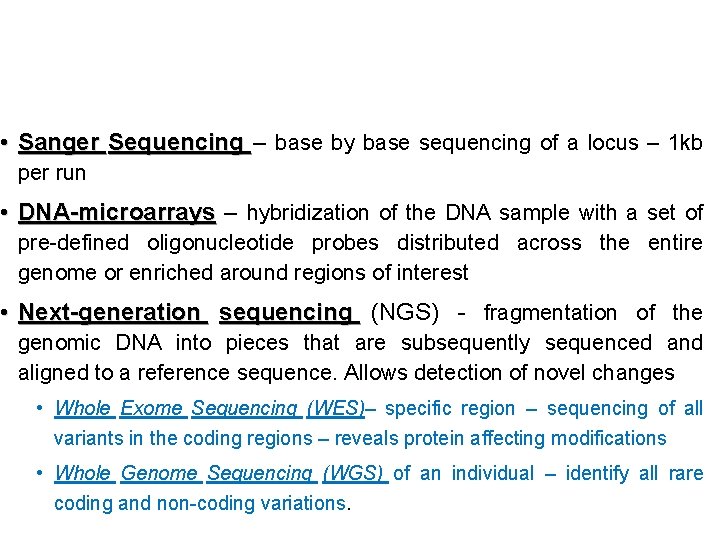
• Sanger Sequencing – base by base sequencing of a locus – 1 kb per run • DNA-microarrays – hybridization of the DNA sample with a set of pre-defined oligonucleotide probes distributed across the entire genome or enriched around regions of interest • Next-generation sequencing (NGS) - fragmentation of the genomic DNA into pieces that are subsequently sequenced and aligned to a reference sequence. Allows detection of novel changes • Whole Exome Sequencing (WES)– specific region – sequencing of all variants in the coding regions – reveals protein affecting modifications • Whole Genome Sequencing (WGS) of an individual – identify all rare coding and non-coding variations.

From: Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences Brief Bioinform. 2016; 19(2): 286 -302. doi: 10. 1093/bib/bbw 114

Questions · Is DNA the only genetic material? · Is DNA sequence/gene alone responsible for heritable traits? · Can heritable changes take place without changes in DNA Sequence?

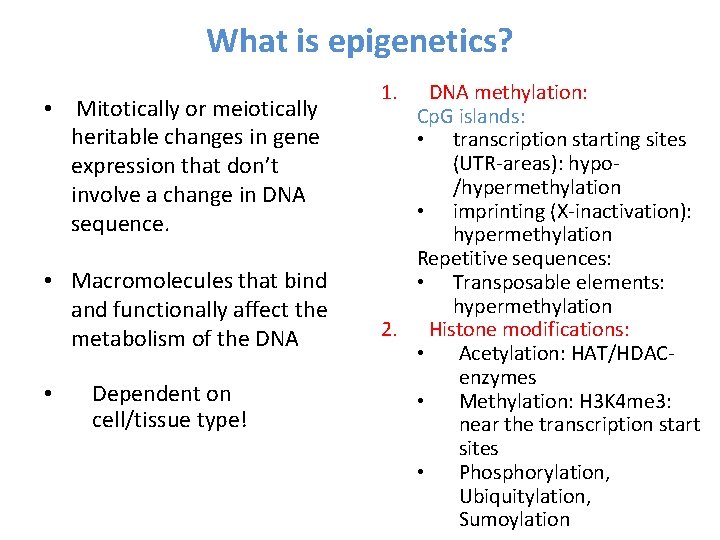
What is epigenetics? • Mitotically or meiotically heritable changes in gene expression that don’t involve a change in DNA sequence. • Macromolecules that bind and functionally affect the metabolism of the DNA • Dependent on cell/tissue type! 1. DNA methylation: Cp. G islands: • transcription starting sites (UTR-areas): hypo/hypermethylation • imprinting (X-inactivation): hypermethylation Repetitive sequences: • Transposable elements: hypermethylation 2. Histone modifications: • Acetylation: HAT/HDAC- enzymes • Methylation: H 3 K 4 me 3: near the transcription start sites • Phosphorylation, Ubiquitylation, Sumoylation

Epigenomics – methylation and histone modification Aug 31 st 2018 Dr. Jacob Junco

Epigenomics • The Encyclopedia of DNA Elements (ENCODE) Program that screens the entire genome and map every detectable functional unit in the database. • identify signature patterns, such as DNA methylation, histone modification and transcription factors, suppressors that bind the specific region. • 88% of trait associated variants detected with GWAS fall in non-coding regions, ENCODE will tremendously impact their assessment and phenotype-related interpretation

DNA Repair, Genomic Instability o DNA is constantly mutating · · · Reactive oxygen species Replication errors Radiation Mutagen Virus o Mutation in a DNA repair gene can cripple the repair process (Werner, Rothmund Thomson) o Repair mechanisms restore DNA to its original state • Application: ü Disease pathology (Cancer, accelerated aging etc) DNA repair, recombination and genomic instability Sept 7 th 2018 Dr. Devon Marie Fitzgerald

Genome editing • Manipulating DNA molecule – study effect of genes – develop novel medicine • Recent advances – rather than studying the DNA taken out of the genome –able to modify genes directly in their endogenous context – any organism • Elucidate their effect in relevant environment – functional organization of the genome at the system levels – identify causal genetic variations. A new era in molecular biology Just CRISPR it!!!

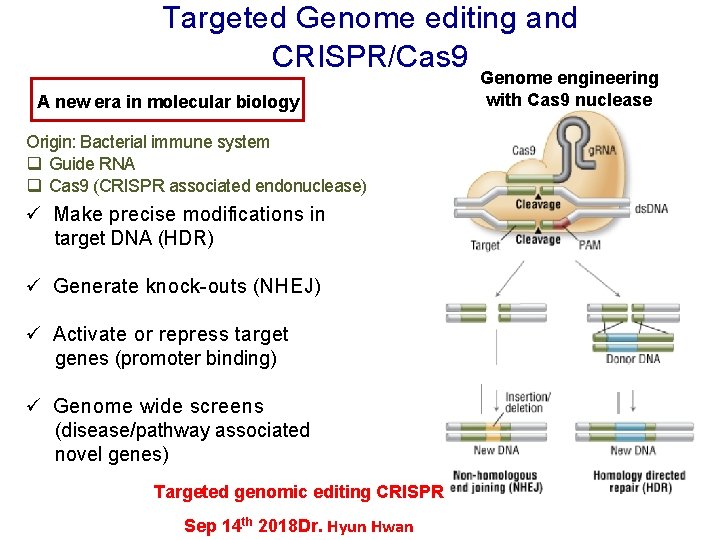
Targeted Genome editing and CRISPR/Cas 9 A new era in molecular biology Origin: Bacterial immune system q Guide RNA q Cas 9 (CRISPR associated endonuclease) ü Make precise modifications in target DNA (HDR) ü Generate knock-outs (NHEJ) ü Activate or repress target genes (promoter binding) ü Genome wide screens (disease/pathway associated novel genes) Targeted genomic editing CRISPR Sep 14 th 2018 Dr. Hyun Hwan Genome engineering with Cas 9 nuclease

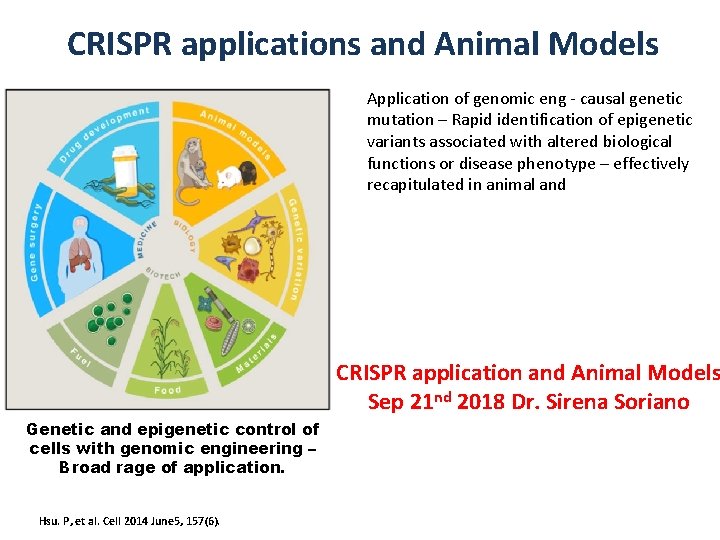
CRISPR applications and Animal Models Application of genomic eng - causal genetic mutation – Rapid identification of epigenetic variants associated with altered biological functions or disease phenotype – effectively recapitulated in animal and CRISPR application and Animal Models Sep 21 nd 2018 Dr. Sirena Soriano Genetic and epigenetic control of cells with genomic engineering – Broad rage of application. Hsu. P, et al. Cell 2014 June 5, 157(6).

Gene Expression: Transcription, Alternative splicing, RNA Seq Regulation of gene expression is the critical link between the genome and cellular morphology/physiology • Transcriptional Factors • Transcriptional Regulation Ø 90% of human genes are alternatively spliced! Sep 28 th 2018 Dr. Tao Ling

Gene Expression: Non-Coding RNA and Personalized Medicine • A non-coding RNA (nc. RNA) is a functional RNA molecule that is not translated into a protein. • Non-coding RNAs: Junk or Critical Regulators in Health and Disease? o Prader–Willi syndrome (sno. RNA-HBII 52) o Alzheimer's disease (lnc. RNA-BACE-AS) o Cancer (mi. R 200) RNAi therapeutics – potent –specific mechanism – targeting gene expression – si. RNA bound o RNA induced silencing complex(RISC) mediated target m. RNA degradation using endonucleases (Arg). Pathway present in all mammalian cells. First-in-Humans Trial of an RNA Interference Therapeutic Targeting VEGF and KSP in Cancer Patients with Liver Involvement Cancer Discovery April 2013 3; 406

Gene expression – Non-coding RNA and personalized medicine. Breaking the genetic enigma Oct – 5 th 2018 – Dr. Narayan Sastri Palla

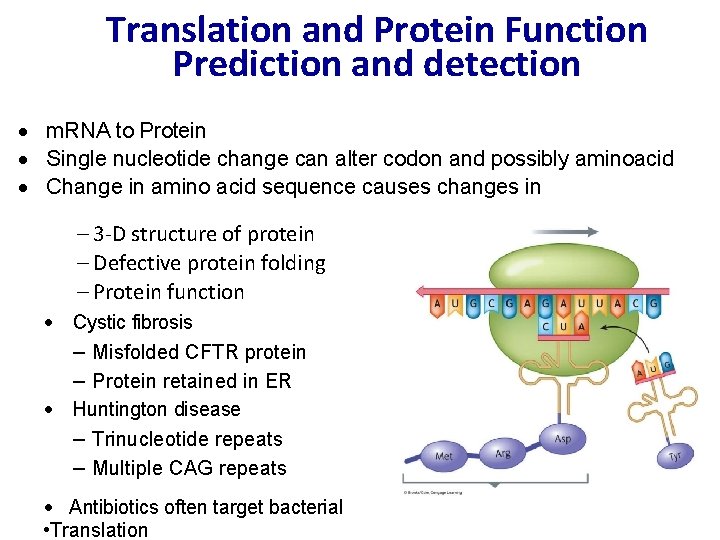
Translation and Protein Function Prediction and detection · m. RNA to Protein · Single nucleotide change can alter codon and possibly aminoacid · Change in amino acid sequence causes changes in – 3 -D structure of protein – Defective protein folding – Protein function · Cystic fibrosis – Misfolded CFTR protein – Protein retained in ER · Huntington disease – Trinucleotide repeats – Multiple CAG repeats · Antibiotics often target bacterial • Translation

From: Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences Brief Bioinform. 2016; 19(2): 286 -302. doi: 10. 1093/bib/bbw 114

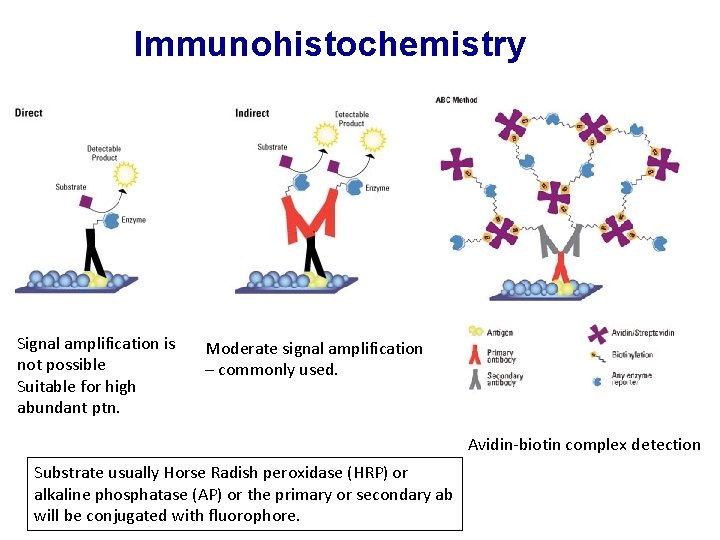
Protein Detection • • Extraction of proteins Separation of proteins Identification of proteins Quantification of Proteins • • • Western Blot Immunohistochemistry/Immunofluorescence ELISA/EIA Immunoprecipitation Mass-Spectrometry X-Ray Chrystallography

• Tissue Preparation for IHC

Immunohistochemistry Signal amplification is not possible Suitable for high abundant ptn. Moderate signal amplification – commonly used. Avidin-biotin complex detection Substrate usually Horse Radish peroxidase (HRP) or alkaline phosphatase (AP) or the primary or secondary ab will be conjugated with fluorophore.

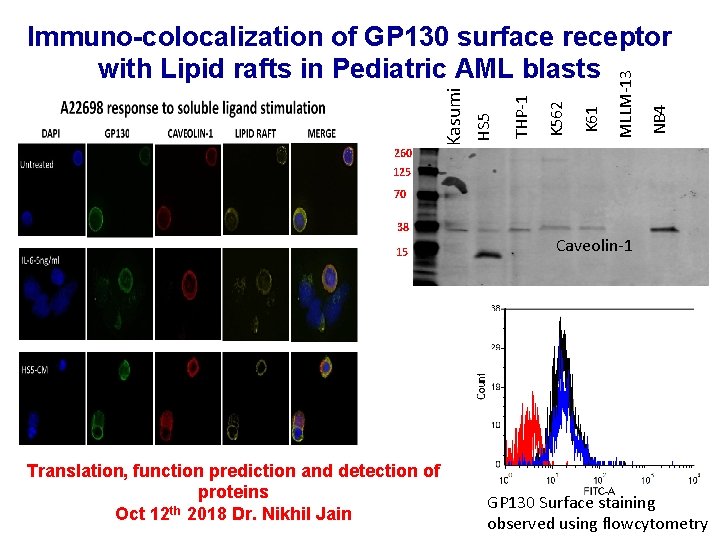
260 NB 4 MLLM-13 K 61 K 562 THP-1 HS 5 Kasumi Immuno-colocalization of GP 130 surface receptor with Lipid rafts in Pediatric AML blasts 125 70 38 15 Translation, function prediction and detection of proteins Oct 12 th 2018 Dr. Nikhil Jain Caveolin-1 GP 130 Surface staining observed using flowcytometry

Protein Modification: Localization and activity · · Enzymes, Receptors, Cytokines, Hormones etc. Translation, Transport and activity Metabolic disorders Types of protein modification Molecular Biology Techniques · Activity Assay · Identification of Post-translational modification Protein Modification – Localization and activity Oct 19 th 2018 Dr. Poonam Sarkar

Cell cycle and chromosomal Instability · Historical context · Preservation and propagation of gene associated diseases · Molecular biology techniques used to these disorders · Molecular biology application in treatment · Introduction to relevant Bio-informatic tools Oct 26 th 2018 Dr. Renuka Ramankutty Menon

Molecular Cloning and its application • Based on isolation of DNA sequence of interest to obtain multiple copies invitro • Insertion of genetic information in to replicating vehicles • Plasmids, viral vectors, BAC etc. • Different techniques of cloning and vectors tailored to the final application. • Lecture – Structure of plasmids, cloning sites, restriction enzymes- cloning softwares and clone a gene in a vector. Nov 2 nd 2018 Dr. Paramahamsa Maturu

Genome-wide association study • A genome-wide association study is an approach that involves rapidly scanning markers across the complete sets of DNA, or genomes, of many people to find genetic variations associated with a particular disease. • Once new genetic associations are identified, researchers can use the information to develop better strategies to detect, treat and prevent the disease. Such studies are particularly useful in finding genetic variations that contribute to common, complex diseases, such as asthma, cancer, diabetes, heart disease and mental illnesses. http: //www. genome. gov/20019523

GWAS- Population Genetics • Introduction to genome-wide association studies (GWAS), clinical relevance of GWAS of complex disease • Introduction to relevant databases to identify haplotype blocks and performing genetic and epigenetic profiling or susceptibility loci • Experimental techniques to determine underlying mechanisms of susceptibility loci • Walkthrough of a leukemia-specific post-GWAS analysis using Haplo. Reg and Regulome. DB •

Potentials of GWAS population Genetics Nov 9 th 2018 Dr. Vince Gant

So by the end of this series …. .

• Bioinformatics · OMIM · Gene Card · Gene · NCBI Primer Design · UCSC Genome Browser · HPRD · Ex. PASy

• OMIM · Online Mendelian Inheritance in Man® · Compendium of genes & genetic phenotypes · Contain information on all known Mendelian • disorders (~15, 000 genes) · Relationship between phenotype & genotype · Initiated 1960 s by Dr. Victor A. Mc. Kusick · Mc. Kusick-Nathans Institute of Genetic • Medicine, JHU · www. omim. org

• Type gene or disease name to get further details

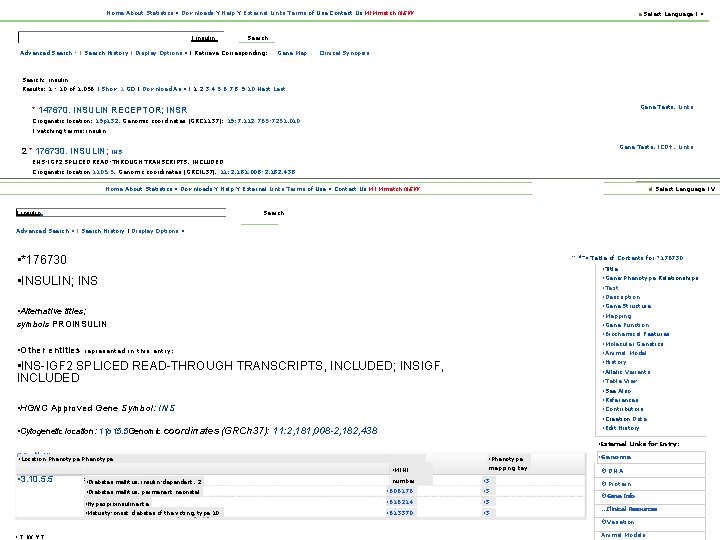
Home About Statistics • Downloads Y Help Y External Links Terms of Use Contact Us MIMmatch NEW a Select Language I • Search I insulin Advanced Search - I Search History I Display Options • I Retrieve Corresponding: Gene Map Clinical Synopsis Search: 'insulin' Results: 1 - 10 of 1, 056 I Show 1 GO I Download As • I 1 2 3 4 5 6 7 8 9 10 Next Last Gene Tests, Links * 147670. INSULIN RECEPTOR; INSR Crogenetic location: 19 p 132, Genomic coordinates (GRC 1137): 19: 7, 112 765 -7251, 010 I'vatching terms: insulin Gene Tests, ICD+, Links 2 * 176730. INSULIN; INS ENS-IGF 2 SPLICED READ-THROUGH TRANSCRIPTS, INCLUDED Crogenetic location 1105. 5, Genomic coordinates (GRCIL 37). 11: 2, 181, 008 -2, 182, 438 Home About Statistics • Downloads Y Help Y External Links Terms of Use • Contact Us MI Mmatch NEW al Select Language I V I insulin Search Advanced Search • I Search History I Display Options • • *176730 • -: 3+ • Table of Contents for *176730 • Title • Gene-Phenotype Relationships • Text • Description • Gene Structure • Mapping • Gene Function • Biochemical Features • Molecular Genetics • Animal Model • History • Allelic Variants • Table View • See Also • References • Contributors • Creation Date • Edit History • INSULIN; INS • Alternative titles; symbols PROINSULIN • Other entities represented in this entry: • INS-IGF 2 SPLICED READ-THROUGH TRANSCRIPTS, INCLUDED; INSIGF, INCLUDED • HGNC Approved Gene Symbol: INS • Cytogenetic location: 11 p 15. 5 Genomic coordinates (GRCh 37): 11: 2, 181, 008 -2, 182, 438 • External Links for Entry: • or. . New. Phenotype • Location • Phenotype • MINI mapping key • 3. 10. 5. 5 • Diabetes mellitus, insulin-dependent, 2 Gene-Phenotype Relationships number • 3 • Diabetes mellitus, permanent neonatal • 606176 125 S 52 • 3 • Hypexproinsulinenta • Maturity-onset diabetes of the voting, type 10 • 616214 • 3 • 613370 • 3 • Genome ·DNA · Protein ·Gene Info. . . Clinical Resources ·Variation • TWYT Animal Models

• Gene · http: //www. ncbi. nlm. nih. gov/gene • F ; 7 2 : • NCBI Resources 1: vi How To NI • Gene No. • Advanced • Help • Gene integrates information from a wide range of species. A record may include nomenclature, Reference Sequences (Ref. Seqs), maps, pathways, variations, phenotypes, and links to genome-, phenotype-, and locus-specific resources worldwide. • Using Gene Tools Other Resources • Gene Quick Start Submit Gene. RIFs 1 -10 molo. Gene • FAQ Submit Correction OMIM • Downiaad. IFTP Statistics Ref. Seo • Ref. Seq Mailing List BLAST Ref. Seq. Gene • Gene News 13 Genome Workbench Uni. Gene • Factsheet Splign Protein Clusters

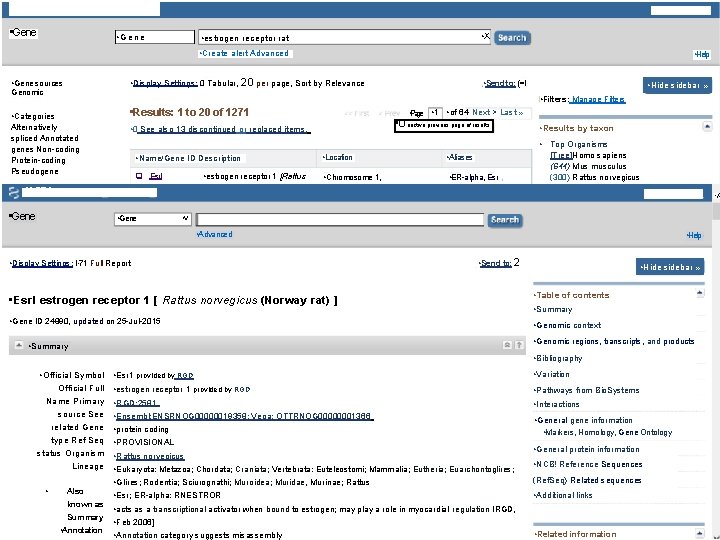
• re, NCBI Resources El How To 17 • Gene • Sign in to NCBI • Gene • X • estrogen receptor rat • Create alert Advanced • Gene sources • Display Settings: 0 Tabular, Genomic • Help 20 per page, Sort by Relevance • Send to: (=I • Hide sidebar » • Filters: Manage Filters • Results: 1 to 20 of 1271 • Categories Alternatively spliced Annotated genes Non-coding Protein-coding Pseudogene • Page • 1 • of 64 Next > Last » • Unactive previous page of results • 0 See also 13 discontinued or replaced items. • Top Organisms • Name/Gene ID Description • Location • Aliases q Esrl • Chromosome 1, • ER-alpha, Esr • estrogen receptor 1 [Rattus • NCBI Resources 17. 1 How To 1, 71 • Gene • Results by taxon • Gene [Tree]Homo sapiens (644) Mus musculus (300) Rattus norvegicus (294) • Sign in to NCBI • V • Advanced • Display Settings: l, 71 Full Report • Help • Send to: 2 • Esrl estrogen receptor 1 [ Rattus norvegicus (Norway rat) ] • Gene ID 24890, updated on 25 -Jul-2015 • Hide sidebar » • Table of contents • Summary • Genomic context • Genomic regions, transcripts, and products • Summary • Bibliography • Official Symbol • Esr 1 provided by RGD Official Full • estrogen receptor 1 provided by RGD Name Primary source See related Gene type Ref Seq status Organism Lineage • Also known as Summary • Annotation • RGD: 2581 • Ensembl: ENSRNOG 00000019358; Vega: OTTRNOG 00000001366 • protein coding • PROVISIONAL • Rattus norvegicus • Eukaryota: Metazoa; Chordata; Craniata; Vertebrata: Euteleostomi; Mammalia; Eutheria; Euarchontoglires; • Glires; Rodentia; Sciurognathi; Muroidea; Muridae, Murinae; Rattus • Esr; ER-alpha: RNESTROR • acts as a transcriptional activator when bound to estrogen; may play a role in myocardial regulation IRGD, • Feb 2006] • Annotation category suggests misassembly • Variation • Pathways from Bio. Systems • Interactions • General gene information • Markers, Homology, Gene Ontology • General protein information • NCB! Reference Sequences (Ref. Seq) Related sequences • Additional links • Related information • A

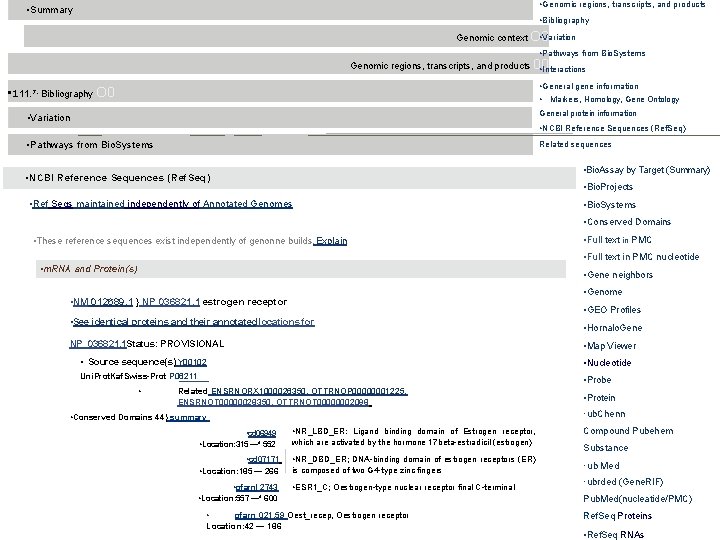
• Genomic regions, transcripts, and products • Summary • Bibliography • Variation Genomic context O 0 • Pathways from Bio. Systems Genomic regions, transcripts, and products 00 • Interactions • General gene information • Markers, Homology, Gene Ontology • 111. 7, Bibliography O 0 • Variation General protein information • Pathways from Bio. Systems Related sequences • NCBI Reference Sequences (Ref. Seq) • Bio. Assay by Target (Summary) • NCBI Reference Sequences (Ref. Seq) • Bio. Projects • Ref Seqs maintained independently of Annotated Genomes • Bio. Systems • Conserved Domains • These reference sequences exist independently of genonne builds. Explain • Full text in PMC nucleotide • m. RNA and Protein(s) • Gene neighbors • Genome • NM 012689. 1 } NP 036821. 1 estrogen receptor • GEO Profiles • See identical proteins and their annotated locations for NP_036821. 1 Status: PROVISIONAL • Hornalo. Gene • Map Viewer • Source sequence(s) Y 00102 • Nucleotide Uni. Prot. Kaf. Swiss-Prot P 06211 • Probe • Related ENSRNORX 1000026350, OTTRNOP 00000001225, ENSRNOT 00000028350, OTTRNOT 00000002098 ·ub. Chenn • Conserved Domains 44) summary • cd 06949 • Location: 315 —* 552 • cd 07171 • Location: 185 — 266 • pfarnl 2743 • NR_LBD_ER: Ligand binding domain of Estrogen receptor, which are activated by the hormone 17 beta-estradicil (estrogen) • NR_DBD_ER; DNA-binding domain of estrogen receptors (ER) is composed of two G 4 -type zinc fingers • ESR 1_C; Oestrogen-type nuclear receptor final C-terminal • Location: 557 —* 600 • • Protein pfarn 021, 59 Oest_recep, Oestrogen receptor Location: 42 — 186 Compound Pubehem Substance · ub. Med ·ubrded (Gene. RIF) Pub. Med(nucleatide/PMC) Ref. Seq Proteins • Ref. Seq RNAs

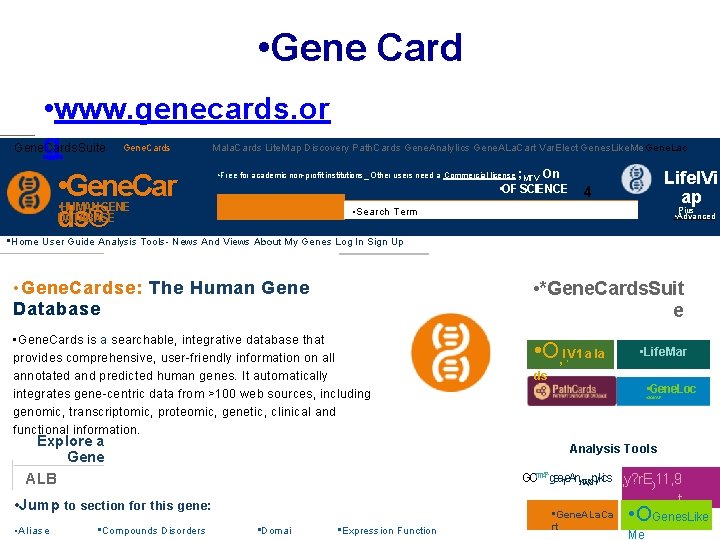
• Gene Card • www. genecards. or Gene. Cards. Suite g • • HUMAN Gene. Car GENE DATABASE ds® • Gene. Cards Mala. Cards Lite. Map Discovery Path. Cards Gene. Analy. Iics Gene. ALa. Cart Var. Elect Genes. Like. Me Gene. Lac • Free for academic non-profit institutions_ Other users need a Commercial license ; MTV • • On • OF SCIENCE Lifel. Vi ap 4 Pius • Search Term • Advanced • Home User Guide Analysis Tools- News And Views About My Genes Log In Sign Up • • Gene. Cardse: The Human Gene Database • *Gene. Cards. Suit e • Gene. Cards is a searchable, integrative database that provides comprehensive, user-friendly information on all annotated and predicted human genes. It automatically integrates gene-centric data from >100 web sources, including genomic, transcriptomic, proteomic, genetic, clinical and functional information. Databases • Life. Mar • O • , Affiliated I V 1 a la Explore a Gene ALB • Jump to section for this gene: • Aliase • Compounds Disorders ds • Gene. Loc • GOICAIE Analysis Tools , GO 111147`gea. Te. An. NW}Spyl kics • Domai • Expression Function • Gene. ALa. Ca rt y? r. E)11, 9 t r • OGenes. Like Me

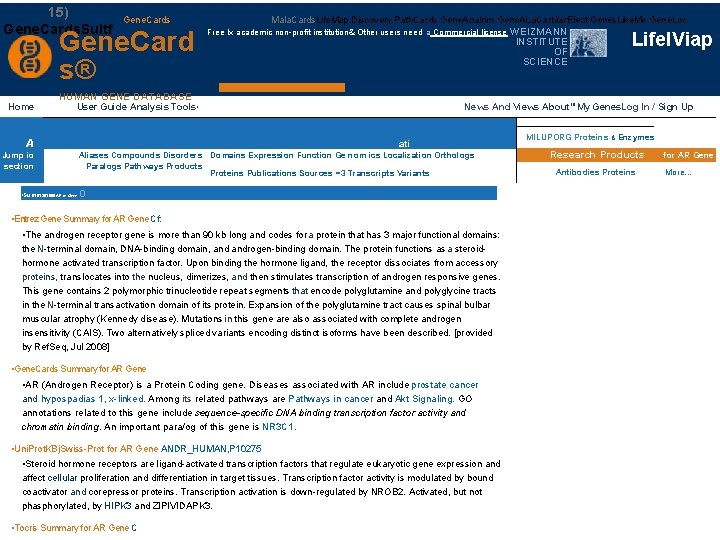
• 15) Gene. Cards. Sultf Home Gene. Cards Gene. Card s® HUMAN GENE DATABASE User Guide Analysis Tools, Mala. Cards. Life. Map Discovery Path. Cards Gene. Ana!rim Gene. ALa. Cart liar. Elect Genes. Like. Me Gene. Loc Free Ix academic non-profit institution& Other users need a Commercial license • Summaries. AR or Gene Lifel. Viap News And Views About"'My Genes. Log In / Sign. Advanced Up ati Aliases Compounds Disorders Domains Expression Function Ge nom ics Localization Orthologs Paralogs Pathways Products Proteins Publications Sources =3 Transcripts Variants o • Entrez Gene Summary for AR Gene Cf: • The androgen receptor gene is more than 90 kb long and codes for a protein that has 3 major functional domains: the N-terminal domain, DNA-binding domain, androgen-binding domain. The protein functions as a steroidhormone activated transcription factor. Upon binding the hormone ligand, the receptor dissociates from accessory proteins, translocates into the nucleus, dimerizes, and then stimulates transcription of androgen responsive genes. This gene contains 2 polymorphic trinucleotide repeat segments that encode polyglutamine and polyglycine tracts in the N-terminal transactivation domain of its protein. Expansion of the polyglutamine tract causes spinal bulbar muscular atrophy (Kennedy disease). Mutations in this gene are also associated with complete androgen insensitivity (CAIS). Two alternatively spliced variants encoding distinct isoforms have been described. [provided by Ref. Seq, Jul 2008] • Gene. Cards Summary for AR Gene • AR (Androgen Receptor) is a Protein Coding gene. Diseases associated with AR include prostate cancer and hypospadias 1, x-linked. Among its related pathways are Pathways in cancer and Akt Signaling. GO annotations related to this gene include sequence-specific DNA binding transcription factor activity and chromatin binding. An important para/og of this gene is NR 3 C 1. • Uni. Prot. KBj. Swiss-Prot for AR Gene ANDR_HUMAN, P 10275 • Steroid hormone receptors are ligand-activated transcription factors that regulate eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Transcription factor activity is modulated by bound coactivator and corepressor proteins. Transcription activation is down-regulated by NROB 2. Activated, but not phasphorylated, by HIPK 3 and ZIPIVIDAPK 3. • Tocris Summary for AR Gene C A Jump io section WEIZMANN INSTITUTE OF SCIENCE MILUPORG Proteins & Enzymes Research Products Antibodies Proteins for AR Gene More. . .

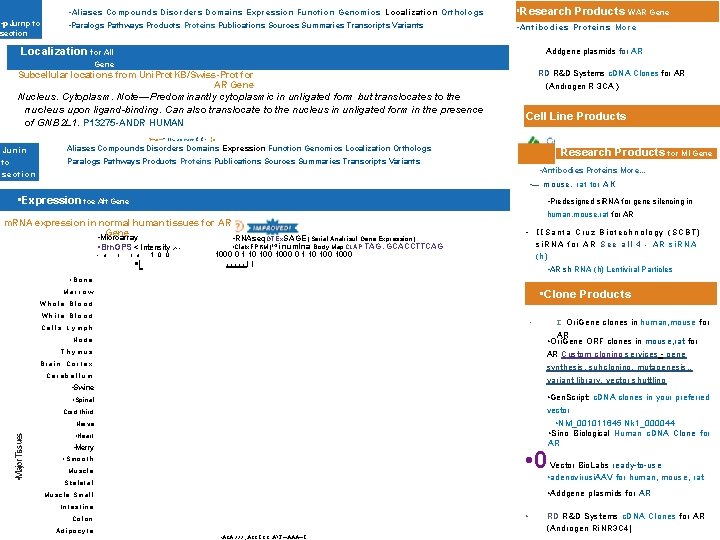
• p. Jurnp to section • Aliases Compounds Disorders Domains Expression Function Genomics Localization Orthologs • Research Products WAR Gene • Paralogs Pathways Products Proteins Publications Sources Summaries Transcripts Variants • Antibodies Proteins More Localization for All Addgene plasmids for AR Gene Subcellular locations from Uni. Prot. KB/Swiss-Prot for AR Gene Nucleus. Cytoplasm. Note—Predominantly cytoplasmic in unligated form but translocates to the nucleus upon ligand-binding. Can also translocate to the nucleus in unligated form in the presence of GNB 2 L 1. P 13275 -ANDR HUMAN I—a: — "—ff. . . an at arr Junin to RD R&D Systems c. DNA Clones for AR (Androgen R 3 CA ) Cell Line Products 1 -1—, (n Aliases Compounds Disorders Domains Expression Function Genomics Localization Orthologs Paralogs Pathways Products Proteins Publications Sources Summaries Transcripts Variants Research Products tor MI Gene • Antibodies Proteins More. . . section • — mouse, rat tor AK • Expression toe Aft Gene • Predesigned si. RNA for gene silencing in human, mouse, rat for AR m. RNA expression in normal human tissues for AR Gene • Microarray • RNAseq GTEx SAGE (Serial Analrisuf Gene Expression) • Cla. Ix. FPKM)14 inumlna Body Map CLAP TAG. GCACCTTCAG • Brn. GPS < Intensity > • IISanta Cruz Biotechnology (SCBT) si. RNA for AR See all 4 - AR si. RNA (h) 5'1 • 0 1 1 0 • I 1 0 0 • 1000 0 1 10 100 1000 • I I I I • AR sh RNA (h) Lentiviral Particles • Bone • Clone Products Marrow Whole Blood White Blood • Cells Lymph I Node Thymus AR Custom cloning services - gene Brain Cortex synthesis, suhcloning, mutagenesis, . Cerebellum variant library, vector shuttling • Swine • Gen. Script: c. DNA clones in your preferred • Spinal vector Cord third • • Major Tissues Nerve • NM_001011645 Nk 1_000044 • Sino Biological Human c. DNA Clone for • Heart • Merry • 0 • Smooth Muscle AR Vector Bio. Labs ready-to-use • adenovirusi. AAV for human, mouse, rat Skeletal • Addgene plasmids for AR Muscle Small Intestine • Colon Adipocyte Ori. Gene clones in human, mouse for AR • Ori. Gene ORF clones in mouse, rat for • ACA AAA. , ACCI. CC, APJ—AAA—I. RD R&D Systems c. DNA Clones for AR (Androgen Ri. NR 3 C 4)

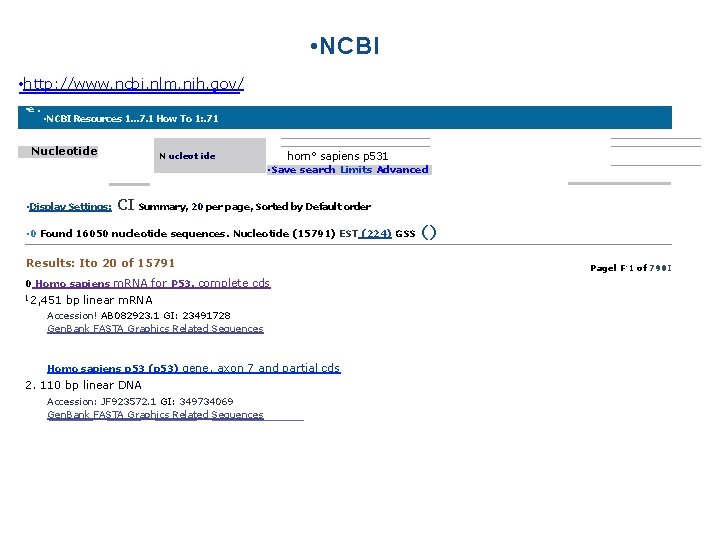
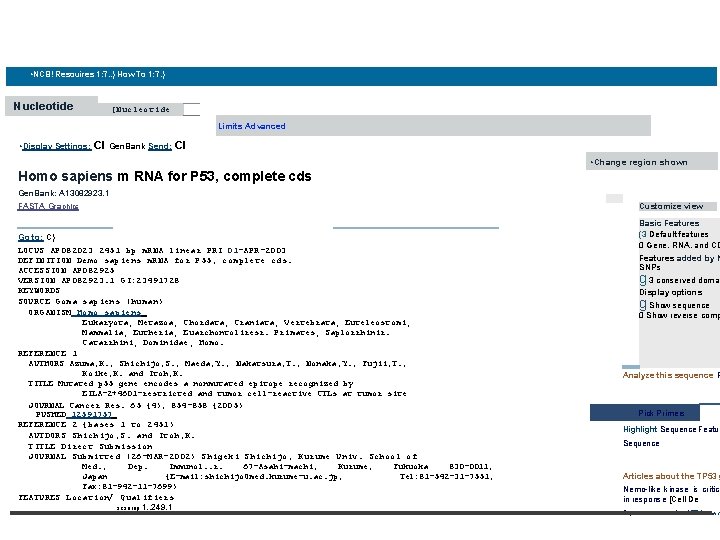
• NCBI • http: //www. ncbi. nlm. nih. gov/ • e. • NCBI Resources 1. . . 7. 1 How To 1: . 71 Nucleotide N ucleot ide horn° sapiens p 531 • Display Settings: • Save search Limits Advanced CI Summary, 20 per page, Sorted by Default order • 0 Found 16050 nucleotide sequences. Nucleotide (15791) EST (224) GSS Results: Ito 20 of 15791 0 Homo sapiens m. RNA for P 53, complete cds 1 2, 451 () Pagel F-1 of 790 I bp linear m. RNA Accession! AB 082923. 1 GI: 23491728 Gen. Bank FASTA Graphics Related Sequences Homo sapiens p 53 (p 53) gene, axon 7 and partial cds 2. 110 bp linear DNA Accession: JF 923572. 1 GI: 349734069 Gen. Bank FASTA Graphics Related Sequences

• NCB! Resouires 1: 7. . ) How To 1: 7. ) Nucleotide [Nucleotide Limits Advanced • Display Settings: CI Gen. Bank Send: CI • Change region shown Homo sapiens m RNA for P 53, complete cds Gen. Bank: A 13082923. 1 FASTA Graphics Basic Features (3 Default features 0 Gene. RNA. and CD Go to: C) LOCUS AP 082023 2451 bp m. RNA linear PRI 01 -APR-2003 DEFINITION Demo sapiens m. RNA for P 55, complete cds. ACCESSION AP 082925 VERSION AP 082923. 1 GI: 23491728 KEYWORDS SOURCE Goma sapiens (human) ORGANISM Homo sapiens Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoliresr. Primates; Saplorrhinir. Catarrhini; Dominidae; Homo. REFERENCE 1 AUTHORS Azuma, K. , Shichijo, S. , Maeda, Y. , Nakatsura, T. , Nonaka, Y. , Fujii, T. , Koike, K. and Itoh, K. TITLE Mutated p 55 gene encodes a nonmutated epitope recognized by EILA-2+4601 -restricted and tumor cell-reactive CTLs at tumor site JOURNAL Cancer Res. 65 {4), 854 -858 {2005) PUSHED 12591757 REFERENCE 2 {bases 1 to 2451) AUTDORS Shichijo, S. and Itoh, K. TITLE Direct Submission JOURNAL Submitted (26 -MAR-2002) Shigeki Shichijo, Kurume Univ. School of Med. , Dep. Immunol. . r. 67 -Asahi-machi, Kurume, Fukuoka 830 -0011, Japan {E-mail: shichijo. Omed. kurume-u. ac. jp, Tel: 81 -542 -31 -7551, Fax: 81 -942 -11 -7699) FEATURES Location/ Qualifiers scsnrnp 1. . 249. 1 Customize view Features added by N SNPs g 3 conserved domai Display options g Show sequence 0 Show reverse comp Analyze this sequence R Pick Primers Highlight Sequence Featur Sequence Articles about the TP 53 g Nemo-like kinase is critica in response [Cell De Prp. rtrintir: uni. He of TPS: WAW

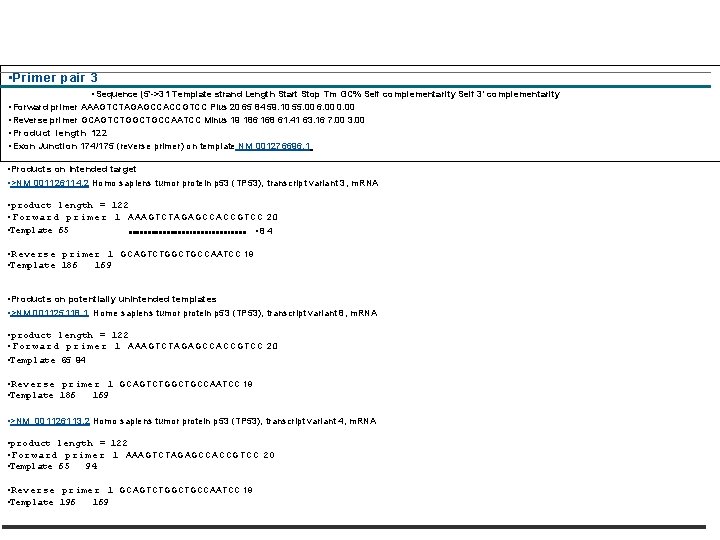
• NCBI Primer Blast • http: //www. ncbi. nlm. nih. gov/tools/primer-blast/ • Primer-BLAST • firictirro • ; 61/Primer-BLAST: Finding primers specific to your PCR template (using Primer 3 and BLAST). More. . . Tips for finding specific primers • Reset page Save search parameters Retrieve recent results • PCR Template • Enter accession, gi, or FASTA sequence (A refseq record is preferred) Clear • Range • AB 082923. 1 • From To • Forward primer • Clem Reverse primer • Or, upload FASTA file • (Choose File j no fib selected • Primer Parameters • Use my own forward primer (5' • Celar ->3' on plus strand) • Use my own reverse primer (5'>3' on minus strand) • Ce l ar • Mln Max • *low- Amplicon size 50 -150 bp for q. PCR • 713 1060 • PCR product size • # of primers to return 0 • 1 • Opt • Mn i • M. Max Tm difference • Primer melting temperatures 57. 0 60. 0 63. 0 3 • (Tm) • Exoniintron selection • A refseq m. RNA sequence as PCR template Input Is required for options In the section • Exon junction span • No preference 1 -41 V. ) Exon junction match Exon at 5' shift Exon at S' s. Ide • 7 '4 • Minimal number of bases that must anneal to exons at the Tor 3' side of the Junction • Intron inclusion • Intron length range q Primer pair must be separated by at least one intron on the =responding genom. Ic DNA 0, • Min Max • 1000 10130 • at a n • a a LIZ a Al

• • Primer pair 3 • Sequence (5'->31 Template strand Length Start Stop Tm GC% Self complementarity Self 3' complementarity • Forward primer AAAGTCTAGAGCCACCGTCC Plus 20 65 84 59. 10 55. 00 6. 00 0. 00 • Reverse primer GCAGTCTGGCTGCCAATCC Minus 19 186 168 61. 41 63. 16 7. 00 3. 00 • Product length 122 • Exon Junction 174/175 (reverse primer) on template NM 001276696. 1 • Products on Intended target • >NM 001126114. 2 Homo sapiens tumor protein p 53 (TP 53), transcript variant 3, m. RNA • product length = 122 • Forward primer 1 A A A G T C T A G C C A C C G T C C 2 0 • Template 65 • 84 • Reverse primer 1 GCAGTCTGGCTGCCAATCC 19 • Template 186 169 • Products on potentially unintended templates • >NM 001125118. 1 Home sapiens tumor protein p 53 (TP 53), transcript variant 8, m. RNA • product length = 122 • Forward primer 1 A A A G T C T A G C C A C C G T C C 2 0 • Template 65 94 • Reverse primer 1 GCAGTCTGGCTGCCAATCC 19 • Template 186 169 • >NM_001126113. 2 Homo sapiens tumor protein p 53 (TP 53), transcript variant 4, m. RNA • product length = 122 • Forward primer 1 AAAGTCTAGAGCCACCGTCC 20 • Template 65 94 • Reverse primer 1 GCAGTCTGGCTGCCAATCC 19 • Template 196 169

• Sequence Similarity Search Tool: • UCSC Browser/BLAT • https: //genome. ucsc. edu/ • UCSC • • Genome Bioinformatics • Genomes - Blat - Tables - Gene Sorter - PCR - Visi. Gene - Session - FAQ - Help • Genome Browser • About the UCSC Genome Bioinformatics Site • Welcome to the UCSC Genome Browser website. This site contains the reference sequence and working draft assemblies fora large collection of • ENCODE genomes. It also provides portals to the ENCODE and Neandertal projects. • Neancle • We encourage you to explore these sequences with ❑urtools. The Genome Browserzoorns and scrolls over chromosomes, showing the work of annotators • Blat • Table • rowser • ne Sorter worldwide. The Gene Sorter shows expression, homology and other information on groups of genes that can be related in many ways. Blat quickly maps your sequence to the genome. The Table Browser provides convenient access to the underlying database. Visi. Gene lets you browse through a large collection of in situ mouse and frog images to examine expression patterns. Genome Graphs allows you to upload and display genome-wide data sets. • The UCSC Genome Browser is developed and maintained by the Genome Bioinformatics Group, a cross-departmental team within the Centerfor Biomolecular Science and Engineering (CBSE) at the University of California Santa Cruz (UCSC) If you have feedback or questions concerning the tools or data on this website, feel free to contact us on ourpublic mailing list • n Silica • R • name The Genome Browser project team relies on public funding to support our work. Donations are welcome — we have many more ideas than ourfunding supports 1 DONATE NOW raphs • laxy • isi. Gene • News D News Archives • To receive announcements of new genome assembly releases, new software features, updates and training seminars by email, subscribe to the genomeannounce mailing list. • Utilities • Down! • lease Log • usto m racks • ncer rowser • 30 July 2014 — New Rat (rni 6) Assembly Now Available in the Genome Browser • We are excited to announce the release of a Genome Browser for the July 2014 assembly of rat, Rattus norvegicus (RGSC Rnor_6. 0, UCSC version rn 6)1 This assembly is provided by the Rat Genome Sequencing Consortium, which is comprised of eight research organizations across the United States and Canada and led by the Baylor College of Medicine. The new RGSC Rnor 6. 0 assembly contains a new, partially assembled Y chromosome as well as improvements to other regions of the genome. You can find more information on the RGSC's efforts to sequence rat genome on the Baylor College of Medicine's project page. • I _I • 2.

• UCSC Browser BLAT • https: //genome. ucsc. edu/ • Genomes Genome Browser Tools Mirrors Downloads My Data Help About Us • Human BLAT Search • • BLAT Search Genome Human Genome: Assembly: 'I 411 [ Dec. 2013 (GRO 138/hg 38) Query type: Sort output Output type: [ DNA query, score'144 hyperlink • AAAGTCTAGAGCCACCCTCC • • • submit) I'm feeling lucky Cclear) • Paste in a query sequence to find its location in the genome. Multiple sequences may be searched if separated by lines starting with '>' followed by the sequence name. • File Upload: Rather than pasting a sequence, you can choose to upload a text file containing the sequence. • Upload sequence: CChoose File) no file selected C submit Me) • Only DNA sequences of 25, 000 or fewer bases and protein ortranslated sequence of 10000 or fewer letters will be processed. Up to 25 sequences can be submitted at the same time. The total limit for multiple sequence submissions is 50, 000 bases or 25, 000 letters. • For locating PCR primers, use In-Silico PCR for best results instead of BLAT.

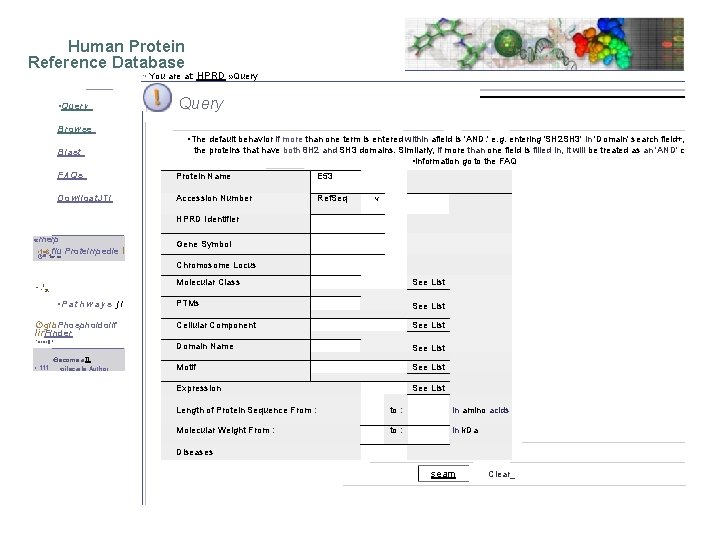
• vvvvw. h p rd. org • Human Protein Reference Database • You are at: HMO • '1 Query 1 04. Browse I oiwas/ • 4 it FAQs • - road • I. • Proteinpedpia • • • EA: • Patirway. S • Phasphti. Volif Findor • fleconie • "Noiccarla Aulhoehly' • News • NOWChn. Okigy • mewl: • "Human Proteinpedia enables data sharing of human proteins" in Februa • bkudinakgy • Phospho. Motif Finder, published in February 2007 issue of nature giotechf • 6. 1 -C • nformatics • Comparison of Protein-Protein Interaction !Databases, published in MC Ei

Human Protein Reference Database • r • Query Browse Blast You are at: HPRD » Query • The default behavior if more than one term is entered within afield is 'AND. ' e. g. entering 'SH 2 SH 3' in 'Domain' search field+, the proteins that have both 8 H 2 and SH 3 domains. Similarly, if more than one field is filled in, it will be treated as an 'AND' c • information go to the FAQs Protein Name E 53 Dowiloat. JTI Accession Number Ref. Seq V HPRD Identifier • is rnerp • 1=6 flu ·. ▪. " • • - • • Proteirrpedie I • . 'n • Pathways ji ·gib. Phosphoidollf lir. Finder • • • • • 1* • Become aill • . . 111 • oilecarle Author Gene Symbol Chromosome Locus Molecular Class See List PTMs See List Cellular Component See List Domain Name See List Motif See List Expression See List Length of Protein Sequence From : to : in amino acids Molecular Weight From : to : in k. Da Diseases seam Clear_

• are at HPRD » Query » p 53 • p 53 ILT: plocular Class Transcription factor • 'Molecular Function Transcription factor ac • Regulation of nucleol • Biological Process nucleoside, nucleotide • acid metabolism ; Apc • GO TO: Isc If ALTERNATE NAMES SUMMARY DISEASES SEQUENCE • PThs&SUBSTRATES • INTERACTIONS ETERNAL LINKS 1 • I Protein Sequence 393 AA NP 0011195341 • HEEPQSDPSI • EPPLSWFS T h. IPLSSSVPSQ RICSDSDGLAP EVRVCACPGR GSRAHSSHLK DLWKLLPENN RTYQGSYGFR LGFLHSGTAK PUIIRVEGN LRVEYLDDRN DRRTEEENLR KKGEPHHELP SKRGINTSRE ECKLMFRTEGP VLSPLPSQAM SVICTYSPAL TFRESVVVPY PGSTRRALPN DS D • - • DDLMLSPDDI • EQWFTEDPGP DEAPRMPEAA PPVAPAPAAP NEMFCQLART TPAAPAPAPS CP 1 NLWVDST PPPGTRVRAM AIYKQSQHHT EPPEVGSDCT EVVRRCPHHE TIHYNYMOINS SCMGGMNRRP ILTIITLEDS NTSSSPQPPY SGNLLGRNSF RPLDGEYFTL QIRGRERFEM FRELNEALEL ZDAQAGREPG

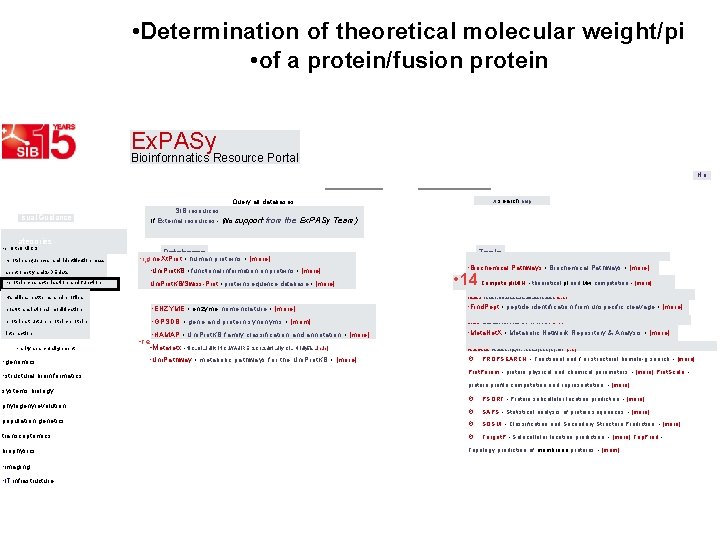
• http: //expasy. org/proteomics • 3 -15 E 11 Ex. PAy • Bioinformatics Resource Portal • IQuery all databases • Visual Guidance • Categories • JI. search help • la SIB resources • If External resources - (No support from the Ex. PASy Teem) • Databases Tools • proteomics • protein sequences and • ne. Xt. Prot • human proteins • [more] identification mass spectrometry and • PROSITE • protein domains and families • [more] 2 -DE data protein characterisation • STRING • protein-protein interactions • [more] and function families, patterns and • 2 SWISS-MODEL Repository • protein structure homology models • [more] • 2 SWISS-MODEL Workspace • structure homology-modeling • [rr Swiss. Dock • protein ligand docking server • [more] · 2 ZIP • Prediction of leucine zipper domains • [more] profiles post-translationa. I • Uni. Prot. KB • functional information on proteins • [more] · 3 of 5 • find user-defined patterns in protein sequences • [more] it modification protein structure • Uni. Prot. KB/Swiss-Prot • protein sequence database • [more] AACompldent • protein identification by as composition • [more] • protein-protein interaction • Viral. Zone • portal to viral Uni. Prot. KB entries • [more] • similarity search/alignment • genomics • structural bioinformatics systems biology phylogenyievolution population genetics transcriptomics biophysics • asy. orgiproteomics • imaging • AAComp. Sim • amino acid composition comparison • [more] • • EMBnet services • bioinformatics tools, databases and courses • [more] • 2 ENZYME • enzyme nomenclature • [more] • GPSDB • gene and protein synonyms • [more] • ria HAMAP • Uni. Prot. KB family classification and annotation • [more] • Meta. Net. X • Metabolic Network Repository & Analysis • [more] • ria MIAPEGel. DB • MIAPE document edition • [more] • ra My. Hits • protein domains database and tools • [more] • PANDITplus • protein families and domains resources • [more] • Pax. Db • protein abundance database • [more] • ria Prolune • Popular science articles (in French) • [more] • Drntoin fulnriol Dnrhal • otnintlir. ol infrornotinn fnr nrntoin • in-inrol • - . - Agadir • Prediction of the helical content of peptides • [more] la ALF • simulation of genome evolution • [more] • I 3 Alignment tools • Four tools for multiple alignments • [more] AIIAII • protein sequences comparisons • [more] · APSSP • Advanced Protein Secondary Structure Prediction • [m • Ascalaph • Molecular modeling software • [more] • fl big-PI • predict GPI modification sites • [more] • 2 Biochemical Pathways • [more] • 2 BLAST • sequence similarity search • [more] • la BLAST (Uni. Prot) • BLAST search on the Uni. Prot web site • [mon • fl BLAST - NCBI • Biological sequence similarity search • [more] • I 5 RI Aq. T _ DPII • PI Ag. T QOUrrh nn nrntoin conlionno ttatalnacoo •

• Determination of theoretical molecular weight/pi • of a protein/fusion protein Ex. PASy Bioinfornnatics Resource Portal Query all databases isual Guidance from the Ex. PASy Team) ategories • pr. Ote 0 Mi. CS • protein sequences and identification mass Databases • r i g ne. Xt. Prot • human proteins • [more] spectrometry and 2 -DE data • Uni. Prot. KB • functional information on proteins • [more] • protein characterisation and function Uni. Prot. KB/Swiss-Prot • protein sequence database • [more] • families, patterns and profiles • ENZYME • enzyme nomenclature • [more] protein structure protein-protein • GPSDB • gene and protein synonyms • [mom] interaction • HAMAP • Uni. Prot. KB family classification and annotation • [more] • rarity search/alignment • structural bioinformatics systems biology Tools • AAComp. Sim • amino acid composition comparison • [more] • Biochemical Pathways • [more] • 14 Compute pli. MIN • theoretical pl and Mw computation • [more] • Find. Mod • protein post-translational modification prediction • [more] post-translational modification • genomics search help SIB resources If External resources - (No support X • re • Meta. Net. X • NICLOILJUIlt. INCLWAUIRE S. C 1. 2 U 6 ILUly CIL rtiridiy 5 is. Linuiej - • Uni. Pathway • metabolic pathways for the Uni. Prot. KB • [more] • Find. Pept • peptide identification from unspecific cleavage • [more] • HAMAP • Uni. Prot. KB family classification and annotation • [more] • Meta. Net. X • Metabolic Network Repository & Analysis • [more] • Predict. Protein - Prediction of physico-chemical protein properties - [more] · PROPSEARCH • Functional and / or structural homolo-g search • [more] Prot. Param • protein physical and chemical parameters • [more] Prot. Scale • protein profile computation and representation • [more] · PSORT • Protein subcellular location prediction • [more] · SAPS • Statistical analysis of protein sequences • [more] · SOSUI • Classification and Secondary Structure Prediction • [more] transcriptomics · Target. P • Sulacellular location prediction • [more] Top. Pred • biophysics Topology prediction of membrane proteins • [mom] phylogenyievolution population genetics • imaging • IT infrastructure Ho

Summary · · · Molecular Biology Processes and Tools Molecular Medicine Bioinformatic tools Data bases

• N-N • ri • I • "Upper panel is my first 4 years of work, • but the lower panel took only 1 year. " Thank you all!! N-N • ihm
 Recent advances in dental ceramics
Recent advances in dental ceramics Payroll in tally
Payroll in tally Short term loans and advances
Short term loans and advances Global oncology trends 2017 advances complexity and cost
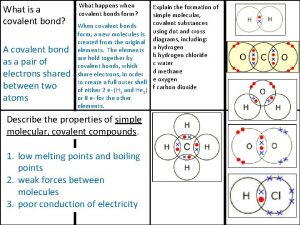
Global oncology trends 2017 advances complexity and cost What is a covalent bond simple definition
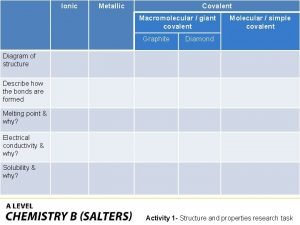
What is a covalent bond simple definition Ionic covalent metallic
Ionic covalent metallic Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Axis powers
Axis powers Advances in technology during wwii
Advances in technology during wwii Lesson 9.1 intellectual advances in the first year
Lesson 9.1 intellectual advances in the first year Advances in real time rendering
Advances in real time rendering Coherent scattering
Coherent scattering Opto-electronic advances
Opto-electronic advances Asset classification norms
Asset classification norms Advances in memory technology
Advances in memory technology Difference between medical report and medical certificate
Difference between medical report and medical certificate Recent trends in ic engine
Recent trends in ic engine Recent developments in ict
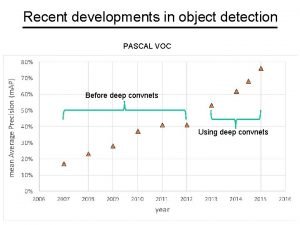
Recent developments in ict Recent developments in object detection
Recent developments in object detection Many recent college graduates have faced
Many recent college graduates have faced Types of international trade
Types of international trade Conclusion of skimming and scanning
Conclusion of skimming and scanning Modern trends in project management
Modern trends in project management Recent demographic changes in the uk
Recent demographic changes in the uk Clever.myips
Clever.myips Stippling
Stippling After kato's serious motorcycle accident
After kato's serious motorcycle accident A friend emails you the results
A friend emails you the results Recent news in passive voice
Recent news in passive voice Https drive google com drive u 0 recent
Https drive google com drive u 0 recent Udin login
Udin login Emerging trends in mis
Emerging trends in mis Mpgu
Mpgu Applications of uv visible spectroscopy
Applications of uv visible spectroscopy Doctors license number
Doctors license number How to start a va medical foster home
How to start a va medical foster home Greater baltimore medical center medical records
Greater baltimore medical center medical records Torrance memorial hospital medical records
Torrance memorial hospital medical records Cartersville medical center medical records
Cartersville medical center medical records Introduction to application layer
Introduction to application layer Introduction to application software
Introduction to application software Introduction to medical informatics
Introduction to medical informatics 2009 delmar cengage learning
2009 delmar cengage learning Bio medical waste introduction
Bio medical waste introduction The word root athr means ________.
The word root athr means ________. Introduction to emergency medical care
Introduction to emergency medical care Introduction to medical virology
Introduction to medical virology Cide medical term
Cide medical term Chapter 1 introduction to medical terminology
Chapter 1 introduction to medical terminology Introduction to medical ethics
Introduction to medical ethics Chapter 1 introduction to medical terminology
Chapter 1 introduction to medical terminology Technologies for biomedical waste treatment
Technologies for biomedical waste treatment Dr neha agrawal
Dr neha agrawal Introduction to emergency medical care
Introduction to emergency medical care Medical
Medical Introduction to emergency medical care
Introduction to emergency medical care Covalent molecular and covalent network
Covalent molecular and covalent network Kinetic molecular theory of solids
Kinetic molecular theory of solids Percentage of composition
Percentage of composition Naming and writing formulas for molecular compounds
Naming and writing formulas for molecular compounds Relationship between bond dipoles and molecular dipoles
Relationship between bond dipoles and molecular dipoles Molecular shape finder
Molecular shape finder Molecular geometry chart
Molecular geometry chart Lewis structure of pf3
Lewis structure of pf3 Molecular geometry and bonding theories
Molecular geometry and bonding theories Electron dot structure of ethene
Electron dot structure of ethene Molecular theory of gases and liquids
Molecular theory of gases and liquids Covalent network solid vs molecular solid
Covalent network solid vs molecular solid