INTRODUCTION AND HISTORY Ribosomes were first observed as


INTRODUCTION AND HISTORY � Ribosomes were first observed as dense particle granules in the mid-1950 s by Romanian cell biologist George Emil Palade using an EM. � Palade in I 974 won a Nobel Prize on this discovery � The term "ribosome" was proposed by Richard B. Roberts in 1958. The ribosome (from ribonucleic acid and the Greek soma, meaning "body") is a large and complex molecular machine, found within all living cells � Venkartraman Ramakrishnan, Thomas A. Steitz and Ada E. Yonath won the Nobel prize in 2009 for determining the detailed molecular structure and mechanism of action of the ribosome. � Ribosome is made of r. RNA and proteins � It serves as the primary site of biological protein synthesis (translation). The cell that have high rate of protein synthesis have large number of ribosome. i. e. human pancrease cells � Cells active in protein synthesis have large nucleoli. � Ribosome link amino acids together by catalyticpeptidyl transferase activity in the order specified by messenger RNA (m. RNA) molecules, thus known ribozyme.
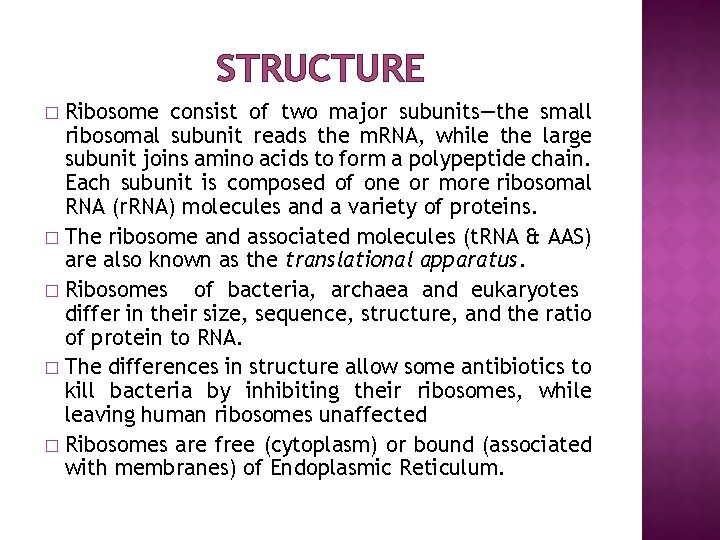
STRUCTURE Ribosome consist of two major subunits—the small ribosomal subunit reads the m. RNA, while the large subunit joins amino acids to form a polypeptide chain. Each subunit is composed of one or more ribosomal RNA (r. RNA) molecules and a variety of proteins. � The ribosome and associated molecules (t. RNA & AAS) are also known as the translational apparatus. � Ribosomes of bacteria, archaea and eukaryotes differ in their size, sequence, structure, and the ratio of protein to RNA. � The differences in structure allow some antibiotics to kill bacteria by inhibiting their ribosomes, while leaving human ribosomes unaffected � Ribosomes are free (cytoplasm) or bound (associated with membranes) of Endoplasmic Reticulum. �
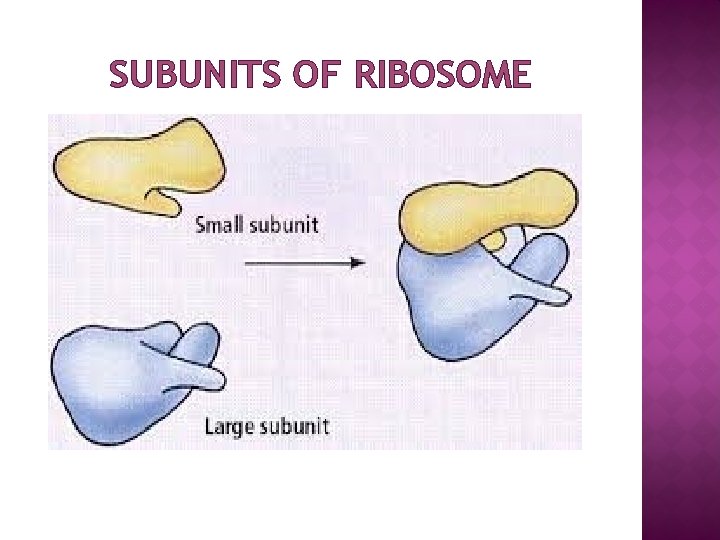
SUBUNITS OF RIBOSOME
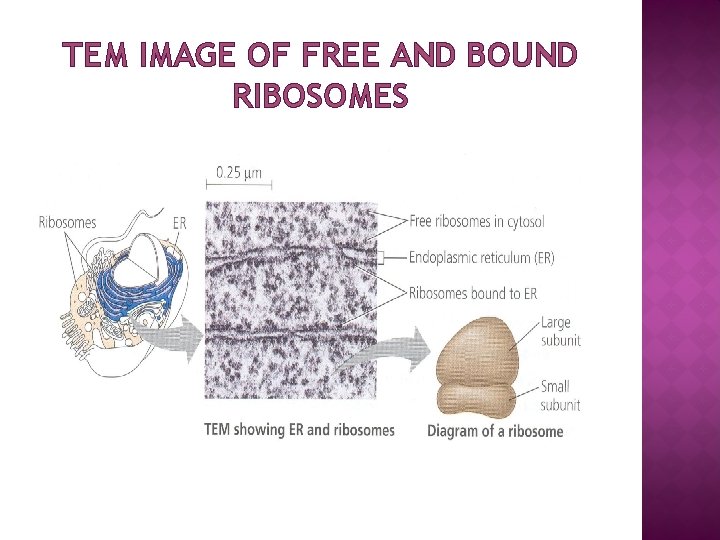
TEM IMAGE OF FREE AND BOUND RIBOSOMES

STRUCTURE OF RIBOSOME
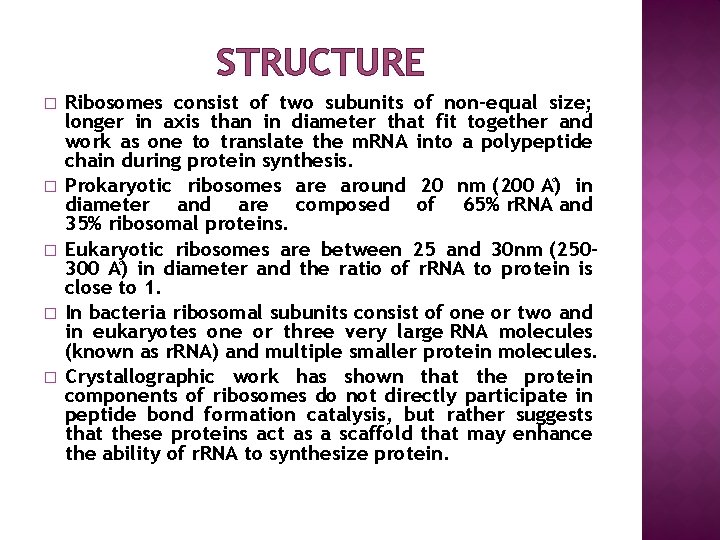
STRUCTURE � � � Ribosomes consist of two subunits of non-equal size; longer in axis than in diameter that fit together and work as one to translate the m. RNA into a polypeptide chain during protein synthesis. Prokaryotic ribosomes are around 20 nm (200 A ) in diameter and are composed of 65% r. RNA and 35% ribosomal proteins. Eukaryotic ribosomes are between 25 and 30 nm (250– 300 A ) in diameter and the ratio of r. RNA to protein is close to 1. In bacteria ribosomal subunits consist of one or two and in eukaryotes one or three very large RNA molecules (known as r. RNA) and multiple smaller protein molecules. Crystallographic work has shown that the protein components of ribosomes do not directly participate in peptide bond formation catalysis, but rather suggests that these proteins act as a scaffold that may enhance the ability of r. RNA to synthesize protein.
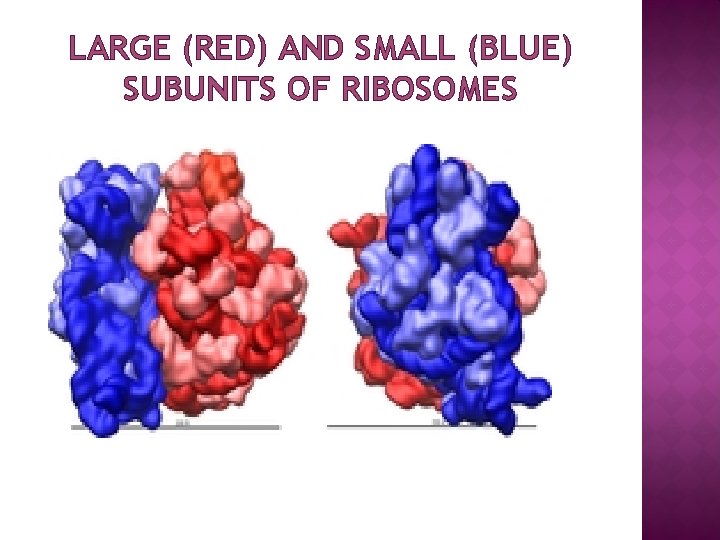
LARGE (RED) AND SMALL (BLUE) SUBUNITS OF RIBOSOMES
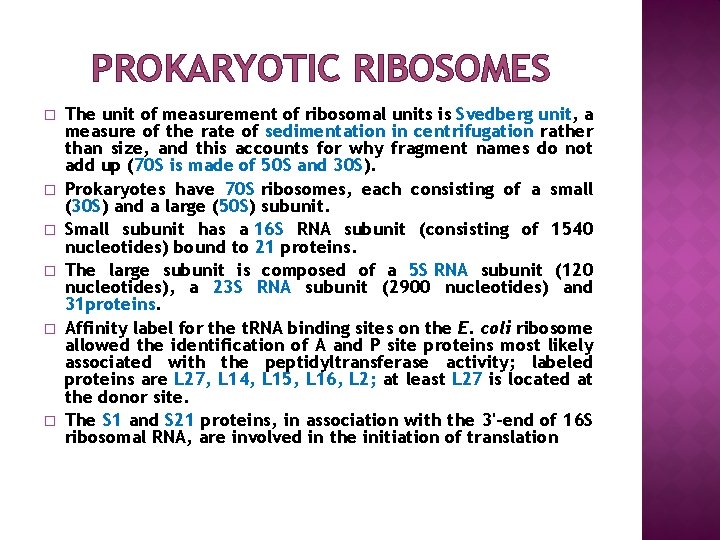
PROKARYOTIC RIBOSOMES � � � The unit of measurement of ribosomal units is Svedberg unit, a measure of the rate of sedimentation in centrifugation rather than size, and this accounts for why fragment names do not add up (70 S is made of 50 S and 30 S). Prokaryotes have 70 S ribosomes, each consisting of a small (30 S) and a large (50 S) subunit. Small subunit has a 16 S RNA subunit (consisting of 1540 nucleotides) bound to 21 proteins. The large subunit is composed of a 5 S RNA subunit (120 nucleotides), a 23 S RNA subunit (2900 nucleotides) and 31 proteins. Affinity label for the t. RNA binding sites on the E. coli ribosome allowed the identification of A and P site proteins most likely associated with the peptidyltransferase activity; labeled proteins are L 27, L 14, L 15, L 16, L 2; at least L 27 is located at the donor site. The S 1 and S 21 proteins, in association with the 3'-end of 16 S ribosomal RNA, are involved in the initiation of translation
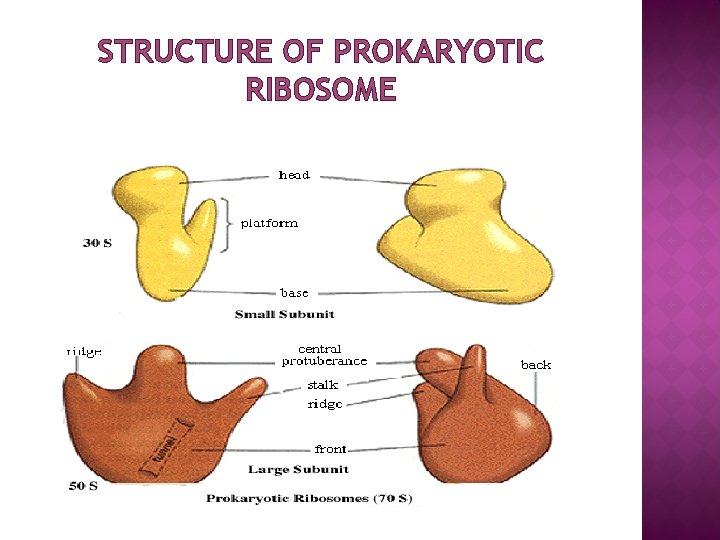
STRUCTURE OF PROKARYOTIC RIBOSOME
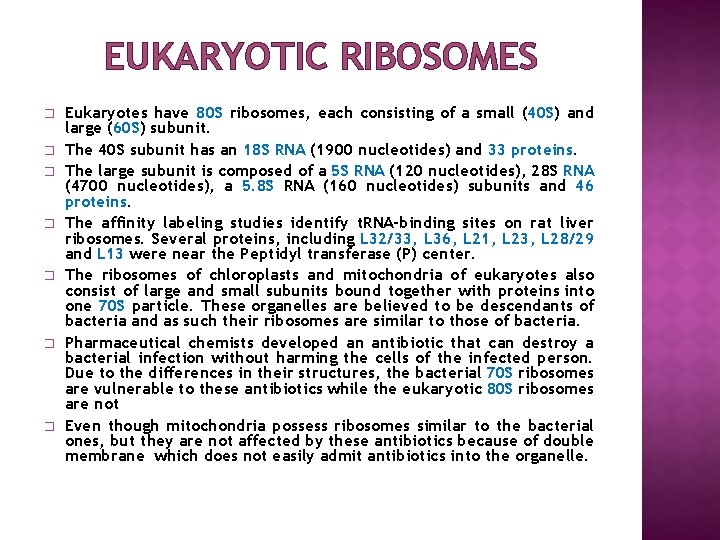
EUKARYOTIC RIBOSOMES � � � � Eukaryotes have 80 S ribosomes, each consisting of a small (40 S) and large (60 S) subunit. The 40 S subunit has an 18 S RNA (1900 nucleotides) and 33 proteins. The large subunit is composed of a 5 S RNA (120 nucleotides), 28 S RNA (4700 nucleotides), a 5. 8 S RNA (160 nucleotides) subunits and 46 proteins. The affinity labeling studies identify t. RNA-binding sites on rat liver ribosomes. Several proteins, including L 32/33, L 36, L 21, L 23, L 28/29 and L 13 were near the Peptidyl transferase (P) center. The ribosomes of chloroplasts and mitochondria of eukaryotes also consist of large and small subunits bound together with proteins into one 70 S particle. These organelles are believed to be descendants of bacteria and as such their ribosomes are similar to those of bacteria. Pharmaceutical chemists developed an antibiotic that can destroy a bacterial infection without harming the cells of the infected person. Due to the differences in their structures, the bacterial 70 S ribosomes are vulnerable to these antibiotics while the eukaryotic 80 S ribosomes are not Even though mitochondria possess ribosomes similar to the bacterial ones, but they are not affected by these antibiotics because of double membrane which does not easily admit antibiotics into the organelle.
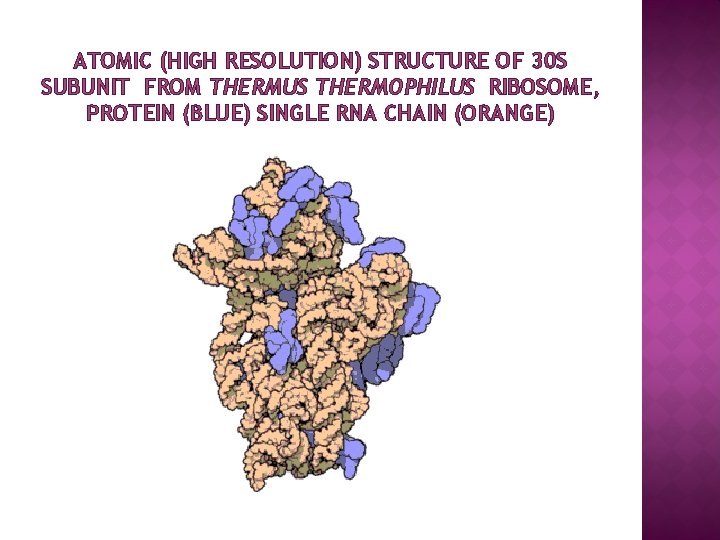
ATOMIC (HIGH RESOLUTION) STRUCTURE OF 30 S SUBUNIT FROM THERMUS THERMOPHILUS RIBOSOME, PROTEIN (BLUE) SINGLE RNA CHAIN (ORANGE)
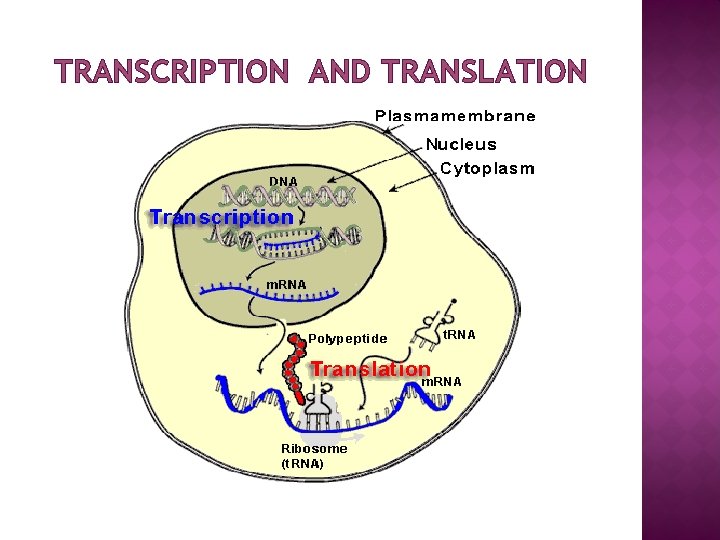
FUNCTION � � � Ribosomes are the workhorses of protein synthesis, the process of translating m. RNA into protein. The m. RNA comprises a series of codons that dictate to the ribosome the sequence of the amino acids needed to make the protein. Using the m. RNA as a template, the ribosome traverses each codon (3 nucleotides) of the m. RNA, pairing it with the appropriate amino acid provided by an aminoacyl-t. RNA. Aminoacyl-t. RNA contains a complementary anticodon on one end and the appropriate amino acid on the other. The small ribosomal subunit, bound to an aminoacyl-t. RNA containing the amino acid methionine, binds to an AUG codon on the m. RNA and recruits the large ribosomal subunit. The ribosome contains three RNA binding sites, designated A, P and E. The A site binds an aminoacyl-t. RNA; the P site binds a peptidyl-t. RNA; and the E site binds a free t. RNA before it exits the ribosome. Protein synthesis begins at a start codon AUG near the 5' end of the m. RNA binds to the P site of the ribosome first. The ribosome is able to identify the start codon by use of the Shine. Dalgarno sequence of the m. RNA in prokaryotes and Kozak box in eukaryotes.
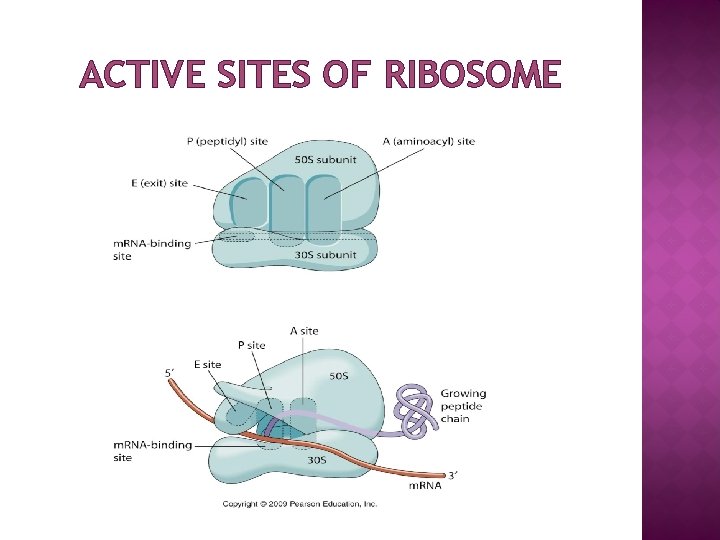
ACTIVE SITES OF RIBOSOME
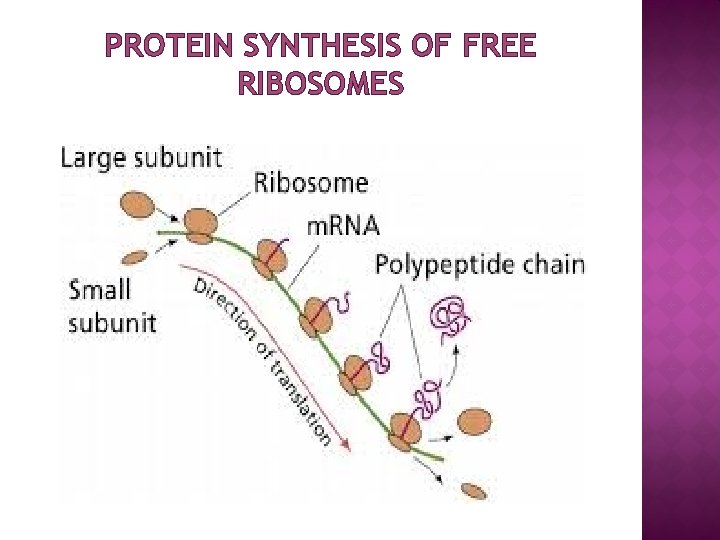
PROTEIN SYNTHESIS OF FREE RIBOSOMES
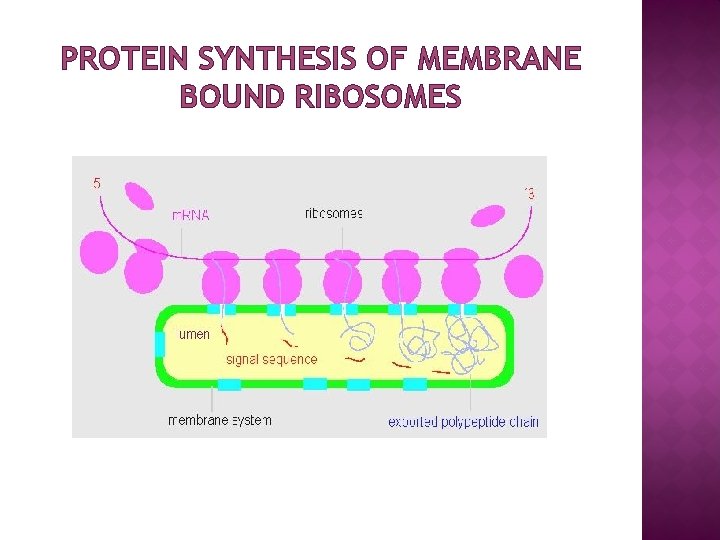
PROTEIN SYNTHESIS OF MEMBRANE BOUND RIBOSOMES
PROTEIN SYNTHESIS AT BOUND RIBOSOMES
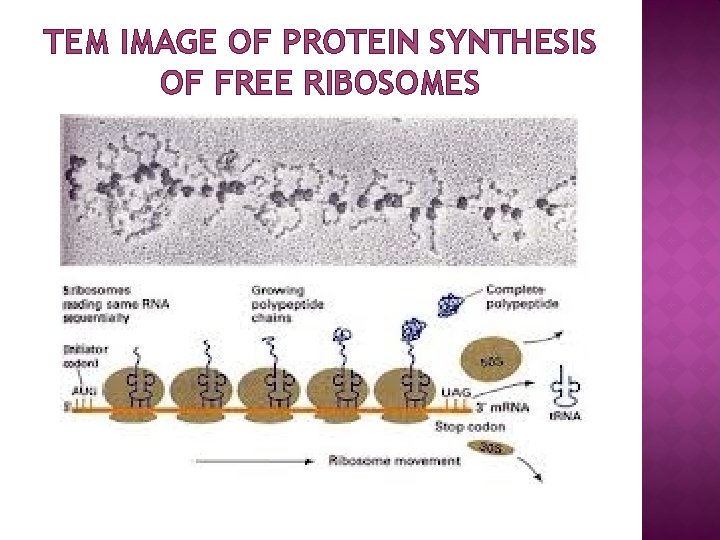
TEM IMAGE OF PROTEIN SYNTHESIS OF FREE RIBOSOMES
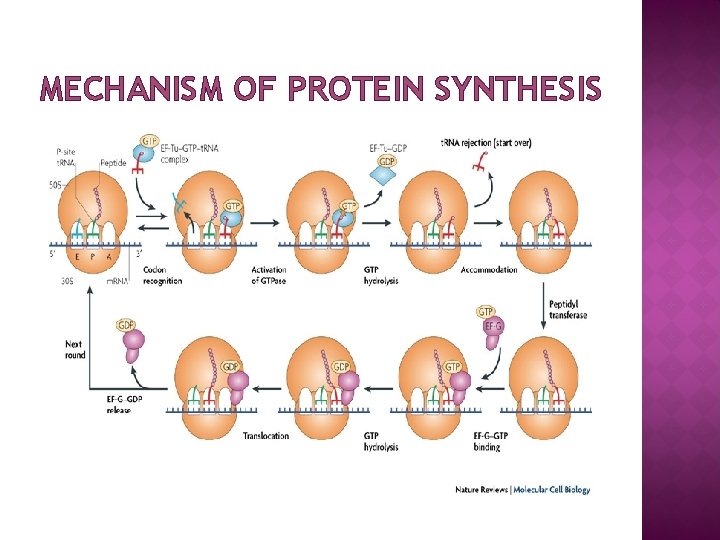
MECHANISM OF PROTEIN SYNTHESIS
LOCATION OF RIBOSOME � � � Ribosomes are of two types: free (cytoplasm) and membrane bound (associated with ER) Free or membrane-bound state depends on the presence of an ER -targeting signal sequence on the protein being synthesized Ribosomes are sometimes referred to as non-membranous organelle. Free ribosomes: Free ribosomes can move about anywhere in the cytosol, but are excluded from the cell nucleus and other organelles. Proteins that are formed from free ribosomes are released into the cytosol and used within the cell. Membrane-bound ribosomes: When a ribosome begins to synthesize proteins that are needed in some organelles, the ribosome making this protein can become "membrane-bound". In eukaryotic cells this happens in a region of the endoplasmic reticulum (ER) called the "rough ER". The newly produced polypeptide chains are inserted directly into the ER by the ribosome undertaking vectorial synthesis and are then transported to their destinations, through the secretory pathway. Bound ribosomes usually produce proteins that are used within the plasma membrane or are expelled from the cell (exocytosis)
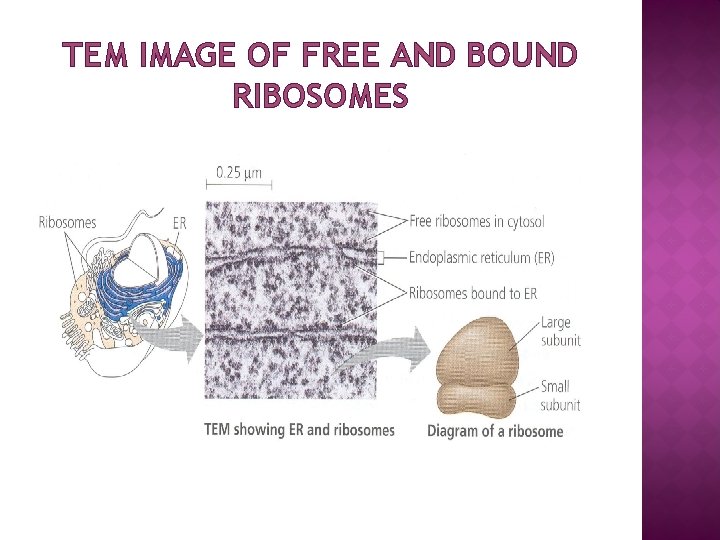
TEM IMAGE OF FREE AND BOUND RIBOSOMES
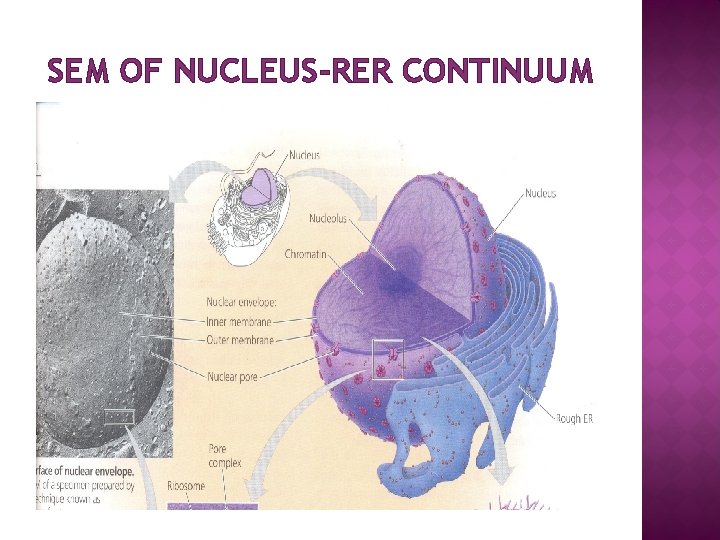
SEM OF NUCLEUS-RER CONTINUUM
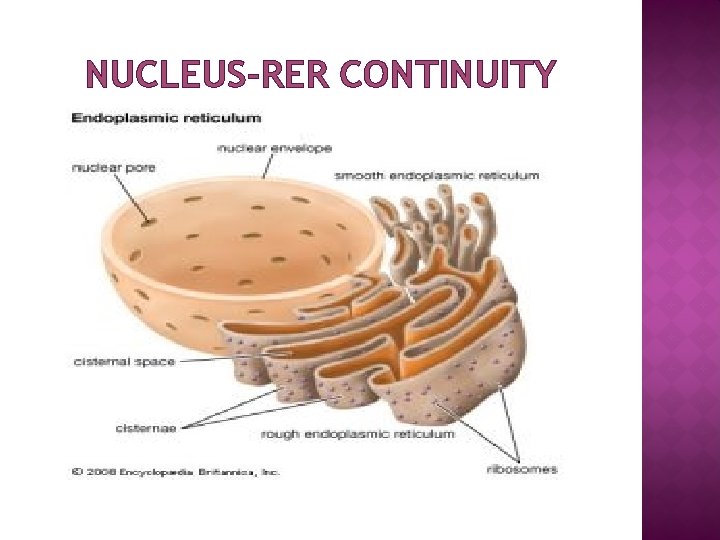
NUCLEUS-RER CONTINUITY
TRANSCRIPTION AND TRANSLATION
- Slides: 24