Intro to molecular dynamics simulation YuShan Lin YSL
- Slides: 22

Intro to molecular dynamics simulation Yu-Shan Lin (YSL) Department of Chemistry Tufts University

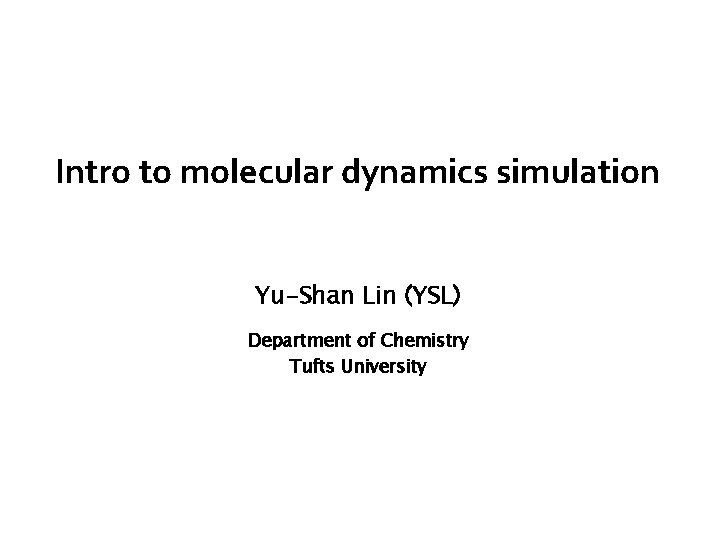
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr s x (0) = 0 m 200 ft m = 1000 kg F = − 7000 N m a = F = − 7 2 s m = 61 m v (t) = v(0)+at = 28− 7 t x ( t) = t 0 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time.

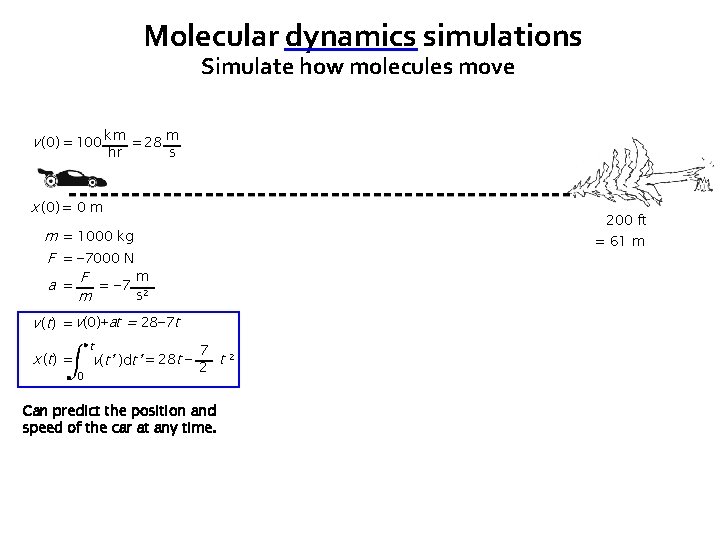
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m v (1 s) = 21 x (0) = 0 m x (1 s) = 24. 5 m hr m s s m = 1000 kg F = − 7000 N m a = F = − 7 2 s m = 61 m v (t) = v(0)+at = 28− 7 t x ( t) = t 0 200 ft 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time.

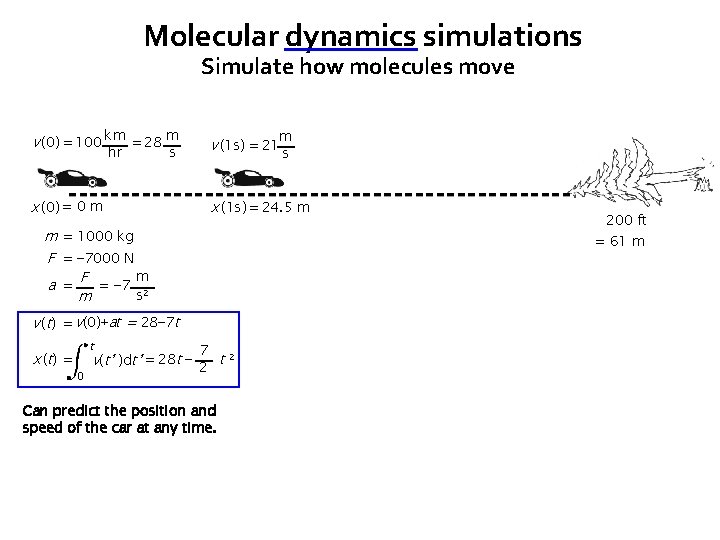
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m v (2 s) = 14 x (0) = 0 m x (2 s) = 42 m hr s m = 1000 kg F = − 7000 N m a = F = − 7 2 s m t 0 200 ft = 61 m v (t) = v(0)+at = 28− 7 t x ( t) = m s 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time.

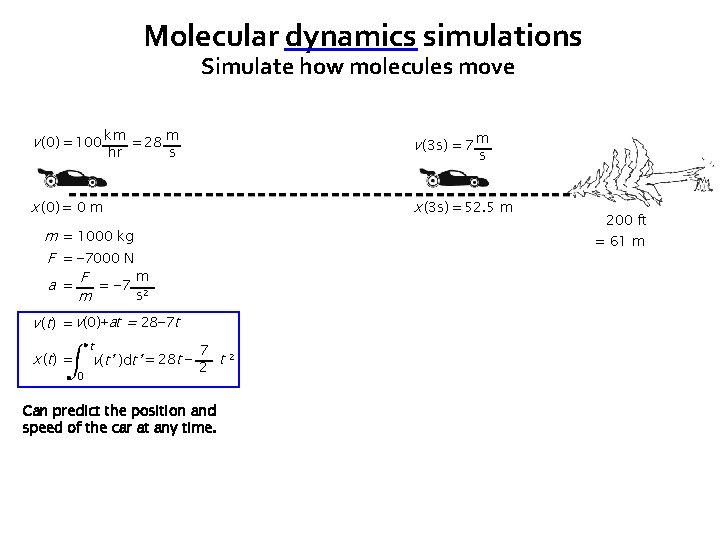
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m v (3 s) = 7 m x (0) = 0 m x (3 s) = 52. 5 m hr s s m = 1000 kg F = − 7000 N m a = F = − 7 2 s m = 61 m v (t) = v(0)+at = 28− 7 t x ( t) = t 0 200 ft 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time.

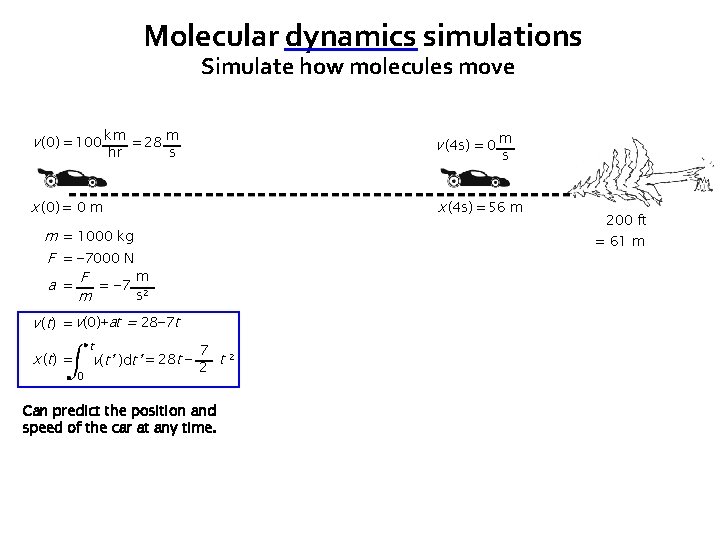
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m v (4 s) = 0 m x (0) = 0 m x (4 s) = 56 m hr s s m = 1000 kg F = − 7000 N m a = F = − 7 2 s m = 61 m v (t) = v(0)+at = 28− 7 t x ( t) = t 0 200 ft 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time.

Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr s x (0) = 0 m m = 1000 kg F = − 7000 N m a = F = − 7 2 s m v (t) = v(0)+at = 28− 7 t x ( t) = t 0 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time.

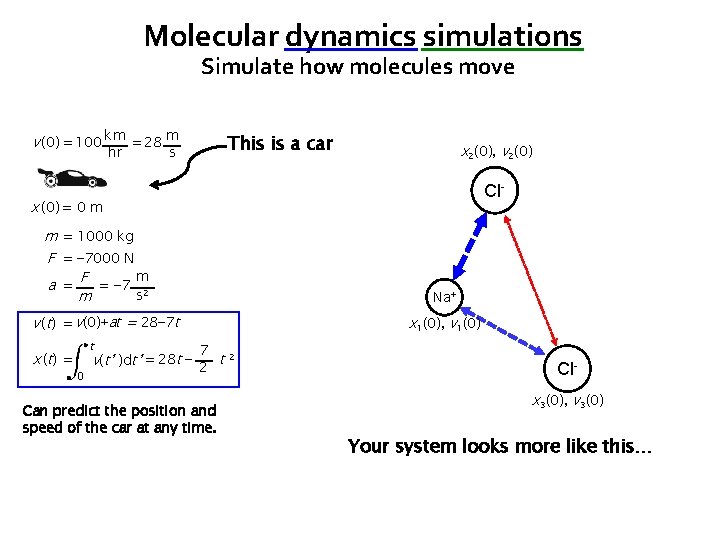
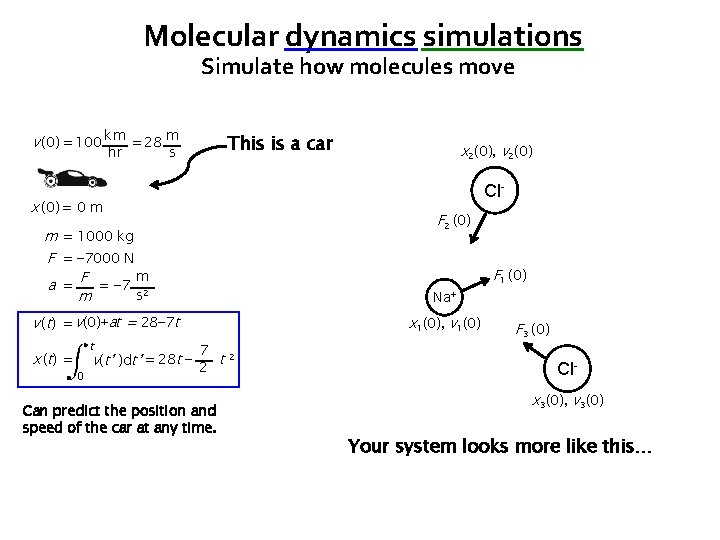
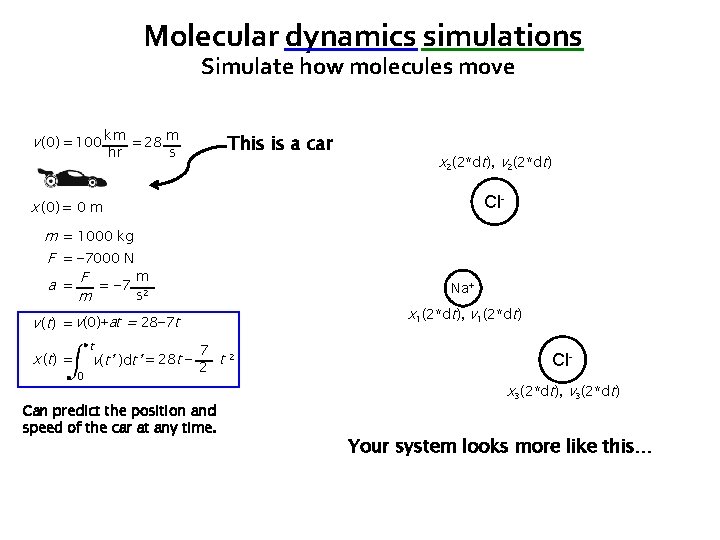
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr This is a car s x 2(0), v 2(0) Cl- x (0) = 0 m m = 1000 kg F = − 7000 N m a = F = − 7 2 s m Na+ v (t) = v(0)+at = 28− 7 t x ( t) = t 0 x 1(0), v 1(0) 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time. Clx 3(0), v 3(0) Your system looks more like this…

Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr This is a car s Cl- x (0) = 0 m F 2 (0) m = 1000 kg F = − 7000 N m a = F = − 7 2 s m F 1 (0) Na+ v (t) = v(0)+at = 28− 7 t x ( t) = t 0 x 2(0), v 2(0) x 1(0), v 1(0) 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time. F 3 (0) Clx 3(0), v 3(0) Your system looks more like this…

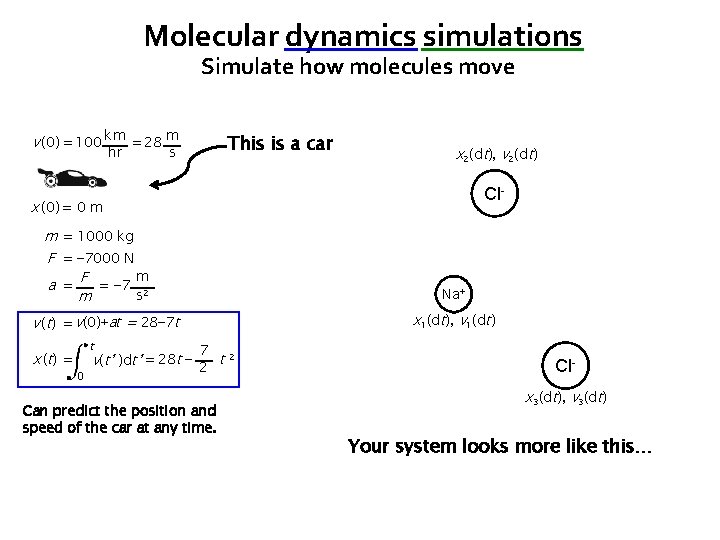
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr This is a car s x 2(dt), v 2(dt) Cl- x (0) = 0 m m = 1000 kg F = − 7000 N m a = F = − 7 2 s m Na+ x 1(dt), v 1(dt) v (t) = v(0)+at = 28− 7 t x ( t) = t 0 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time. Clx 3(dt), v 3(dt) Your system looks more like this…

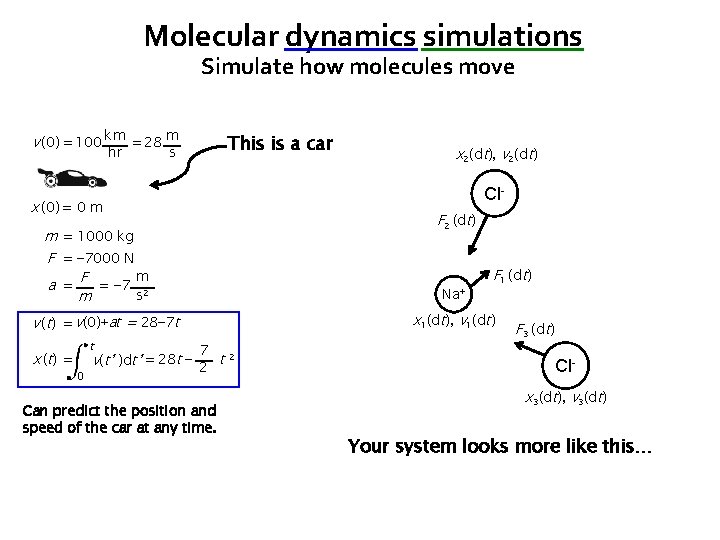
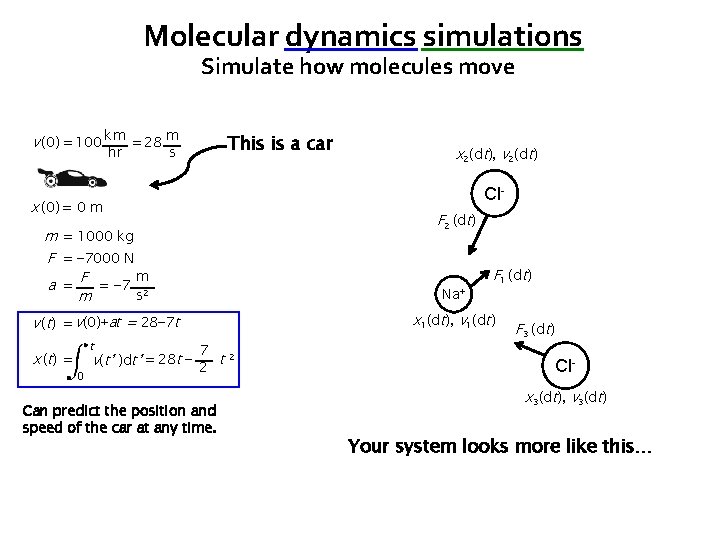
Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr This is a car s Cl- x (0) = 0 m F 2 (dt) m = 1000 kg F = − 7000 N m a = F = − 7 2 s m Na+ t 0 F 1 (dt) x 1(dt), v 1(dt) v (t) = v(0)+at = 28− 7 t x ( t) = x 2(dt), v 2(dt) 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time. F 3 (dt) Clx 3(dt), v 3(dt) Your system looks more like this…

Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr This is a car s Cl- x (0) = 0 m F 2 (dt) m = 1000 kg F = − 7000 N m a = F = − 7 2 s m Na+ t 0 F 1 (dt) x 1(dt), v 1(dt) v (t) = v(0)+at = 28− 7 t x ( t) = x 2(dt), v 2(dt) 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time. F 3 (dt) Clx 3(dt), v 3(dt) Your system looks more like this…

Molecular dynamics simulations Simulate how molecules move v (0) = 100 km = 28 m hr This is a car s x 2(2*dt), v 2(2*dt) Cl- x (0) = 0 m m = 1000 kg F = − 7000 N m a = F = − 7 2 s m Na+ x 1(2*dt), v 1(2*dt) v (t) = v(0)+at = 28− 7 t x ( t) = t 0 7 v(t’ )dt’ = 28 t − 2 t 2 Can predict the position and speed of the car at any time. Clx 3(2*dt), v 3(2*dt) Your system looks more like this…

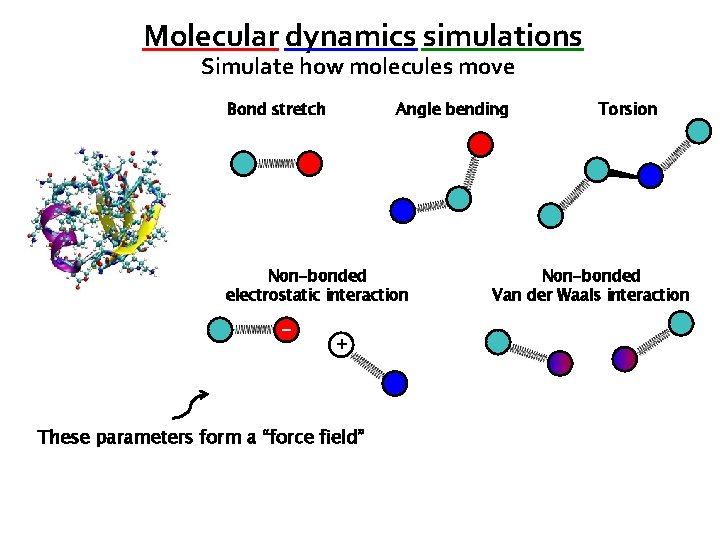
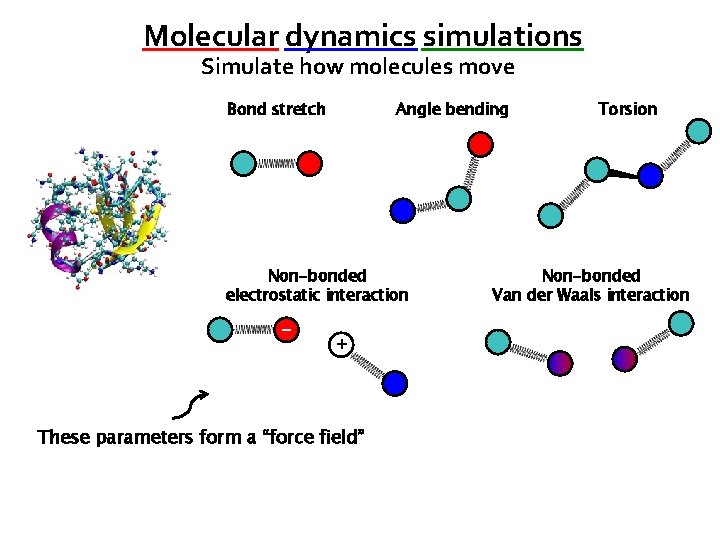
Molecular dynamics simulations Simulate how molecules move Bond stretch Angle bending Non-bonded electrostatic interaction − + These parameters form a “force field” Torsion Non-bonded Van der Waals interaction

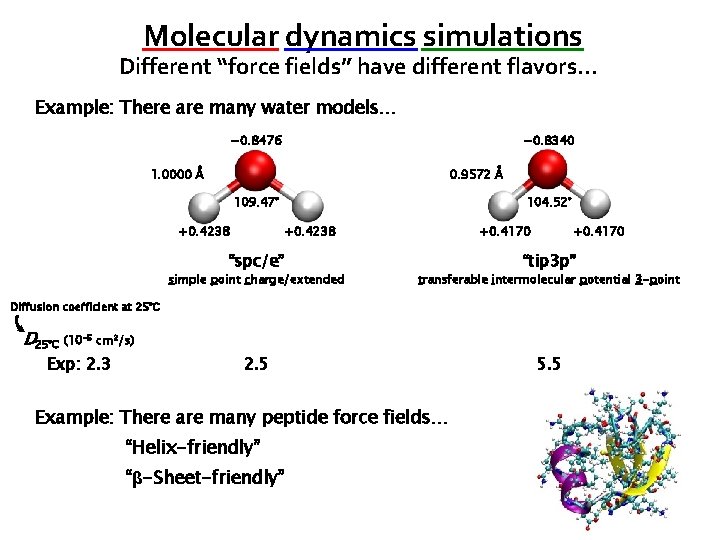
Molecular dynamics simulations Different “force fields” have different flavors… Example: There are many water models… − 0. 8476 − 0. 8340 1. 0000 Å 0. 9572 Å 109. 47° +0. 4238 104. 52° +0. 4238 “spc/e” +0. 4170 “tip 3 p” simple point charge/extended transferable intermolecular potential 3 -point 2. 5 5. 5 Diffusion coefficient at 25°C D 25°C (10 -5 cm 2/s) Exp: 2. 3 Example: There are many peptide force fields… “Helix-friendly” “β-Sheet-friendly”

Molecular dynamics simulations Simulate how molecules move Bond stretch Angle bending Non-bonded electrostatic interaction − + These parameters form a “force field” Torsion Non-bonded Van der Waals interaction

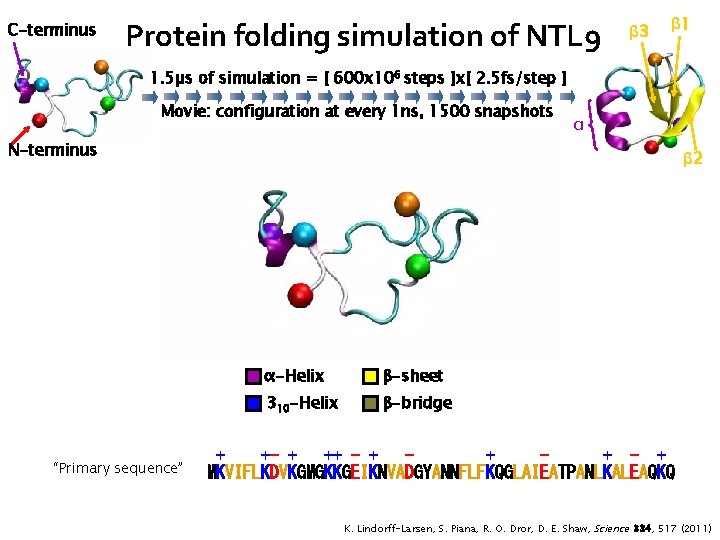
C-terminus Protein folding simulation of NTL 9 β 3 β 1 1. 5µs of simulation = [ 600 x 106 steps ]x[ 2. 5 fs/step ] Movie: configuration at every 1 ns, 1500 snapshots α N-terminus “Primary sequence” β 2 α-Helix β-sheet 310 -Helix β-bridge + +- + ++ - + + + - + MKVIFLKDVKGMGKKGEIKNVADGYANNFLFKQGLAIEATPANLKALEAQKQ K. Lindorff-Larsen, S. Piana, R. O. Dror, D. E. Shaw, Science 334, 517 (2011)

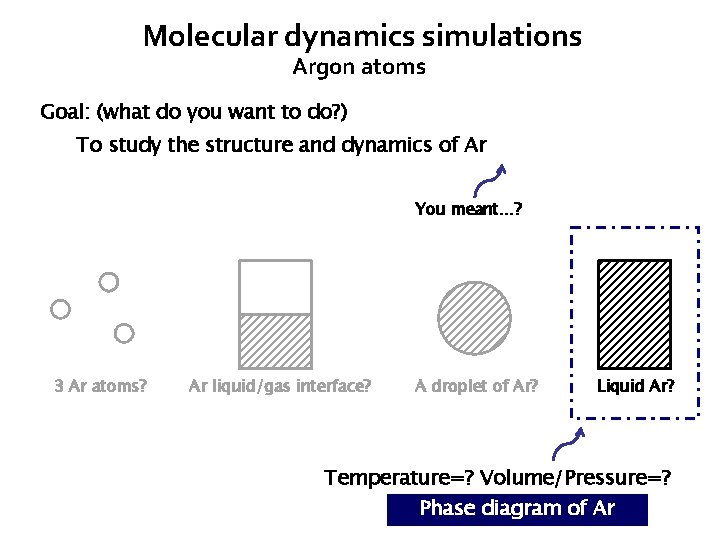
Molecular dynamics simulations Argon atoms Goal: (what do you want to do? ) To study the structure and dynamics of Ar You meant…? 3 Ar atoms? Ar liquid/gas interface? A droplet of Ar? Liquid Ar? Temperature=? Volume/Pressure=? Phase diagram of Ar

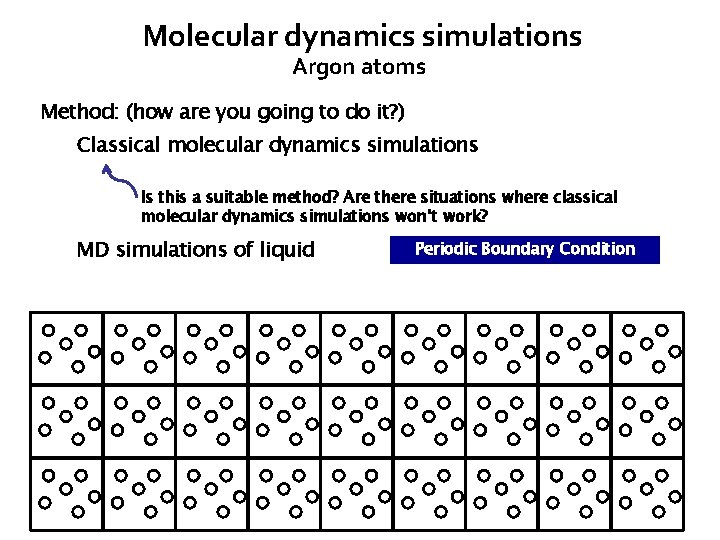
Molecular dynamics simulations Argon atoms Method: (how are you going to do it? ) Classical molecular dynamics simulations Is this a suitable method? Are there situations where classical molecular dynamics simulations won’t work? MD simulations of liquid Periodic Boundary Condition

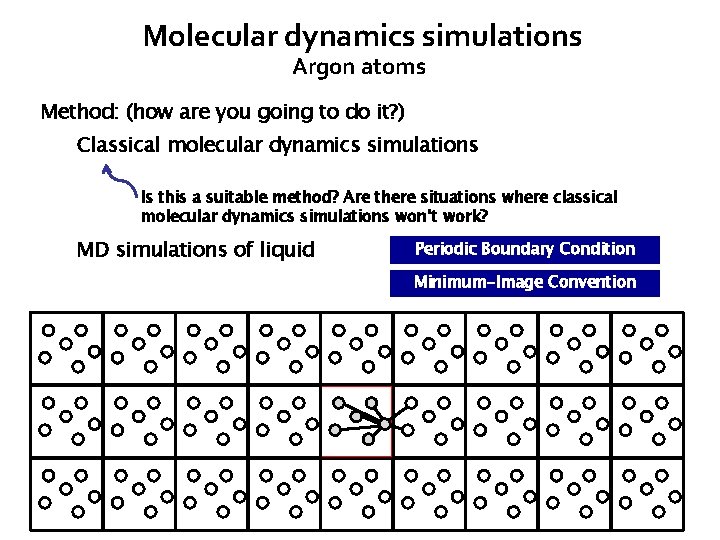
Molecular dynamics simulations Argon atoms Method: (how are you going to do it? ) Classical molecular dynamics simulations Is this a suitable method? Are there situations where classical molecular dynamics simulations won’t work? MD simulations of liquid Periodic Boundary Condition Minimum-Image Convention

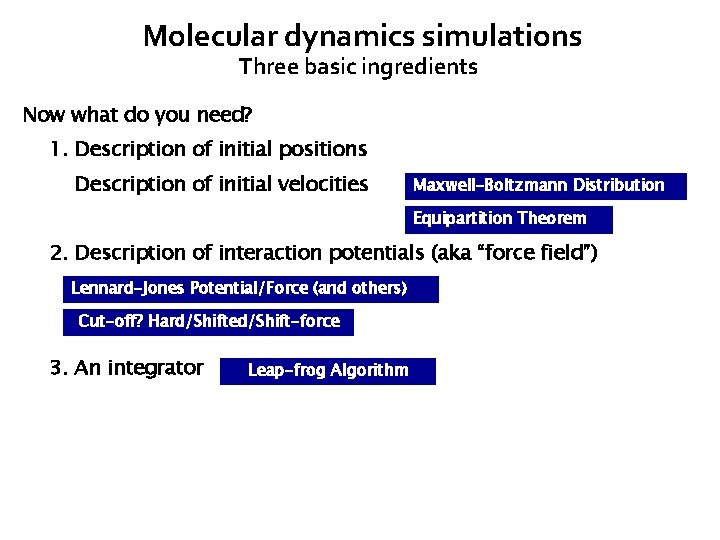
Molecular dynamics simulations Three basic ingredients Now what do you need? 1. Description of initial positions 1. Description of initial velocities Maxwell-Boltzmann Distribution Equipartition Theorem 2. Description of interaction potentials (aka “force field”) Lennard-Jones Potential/Force (and others) Cut-off? Hard/Shifted/Shift-force 3. An integrator Leap-frog Algorithm

Molecular dynamics simulations To learn more about running MD simulations, visit our website at http: //ase. tufts. edu/chemistry/lin/outreach. html