Interpreting Microarray Expression Data Using Text Annotating the









- Slides: 21

Interpreting Microarray Expression Data Using Text Annotating the Genes Michael Molla, Peter Andreae, Jeremy Glasner, Frederick Blattner, Jude Shavlik University of Wisconsin – Madison

The Basic Task Given Microarray Expression Data & Text Annotations of Genes Generate Model of Expression

Motivation • Lots of Data Available on the Internet – Microarray Expression Data – Text Annotations of Genes • Maybe we can Make the Scientist’s Job Easier – Generate a Model of Expression Automatically – Easier First Step for the Human

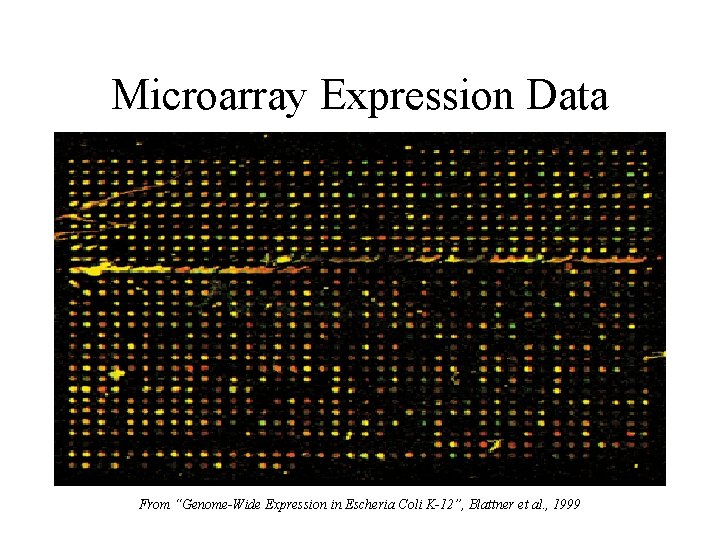
Microarray Expression Data • Each spot represents a gene in E. coli • Colors Indicate Up- or Down-Regulation Under Antibiotic Shock • Four Purpose 3 Classes – Up-Regulated – Down-Regulated – No-Change

Microarray Expression Data From “Genome-Wide Expression in Escheria Coli K-12”, Blattner et al. , 1999

Our Microarray Experiment • • • 4290 genes 574 up-regulated 333 down-regulated 2747 un-regulated 636 non enough signal

Text Annotations of Genes • The text from a sample Swiss. Prot entry (b 1382) – The “description” field HYPOTHETICAL 6. 8 KDA PROTEIN IN LDHA-FEAR INTERGENIC REGION – The “keyword” field HYPOTHETICAL PROTEIN

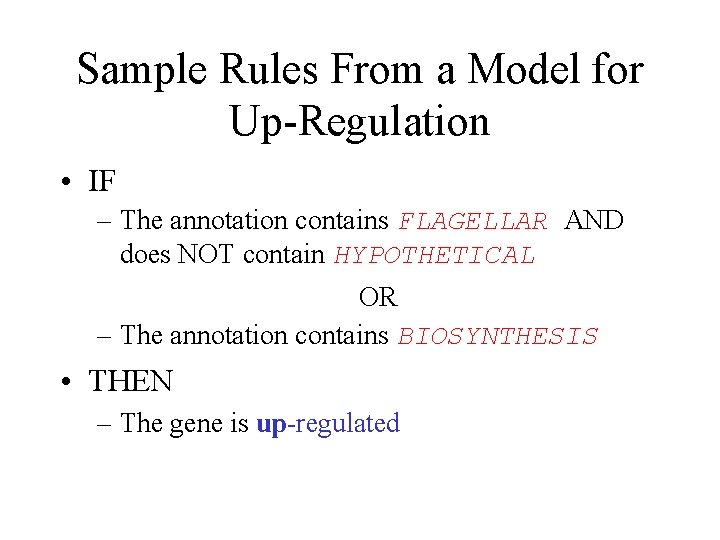
Sample Rules From a Model for Up-Regulation • IF – The annotation contains FLAGELLAR AND does NOT contain HYPOTHETICAL OR – The annotation contains BIOSYNTHESIS • THEN – The gene is up-regulated

Why use Machine Learning? • Concerned with machines learning from available data • Informed by text data, the leaner can make first-pass model for the scientist

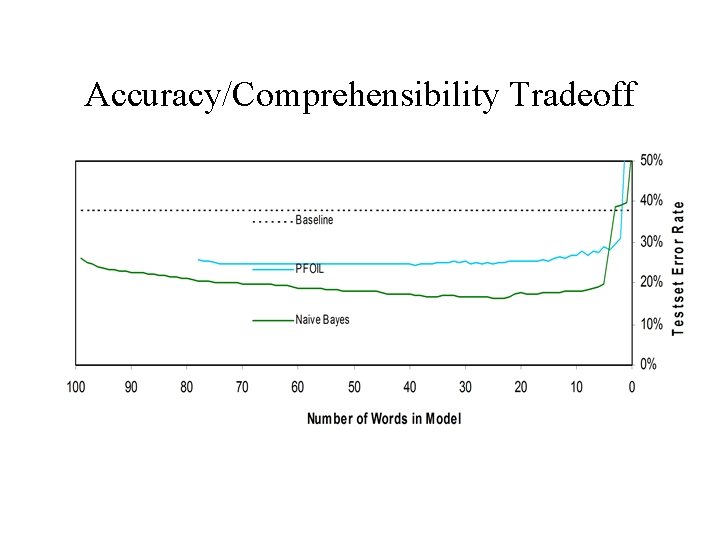
Desired Properties of a Model • Accurate – Measure with cross validation • Comprehensible – Measure with model size • Stable to Small Changes in the Data – Measure with random subsampling

Approaches • Naïve Bayes – Statistical method – Uses all of the words (present or absent) • PFOIL – Covering algorithm – Chooses words to use one at a time

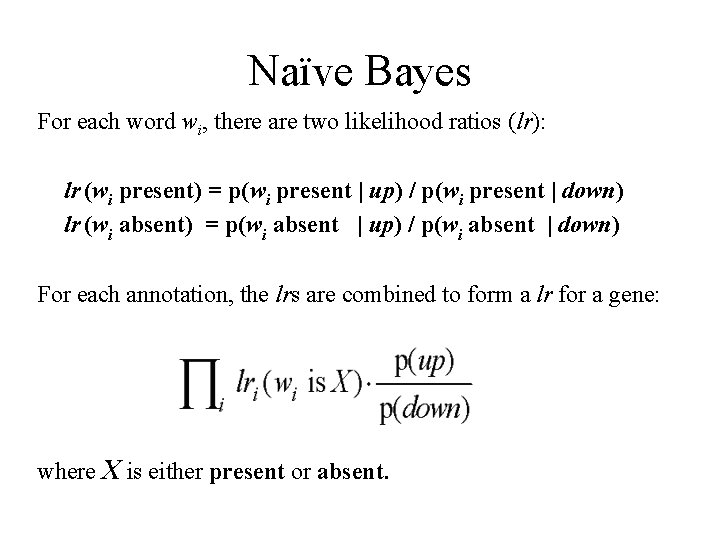
Naïve Bayes For each word wi, there are two likelihood ratios (lr): lr (wi present) = p(wi present | up) / p(wi present | down) lr (wi absent) = p(wi absent | up) / p(wi absent | down) For each annotation, the lrs are combined to form a lr for a gene: where X is either present or absent.

PFOIL • • Learn rules from data Produces multiple if-then rules from data Builds rules by adding one word at a time Easy to interpret models

Accuracy/Comprehensibility Tradeoff

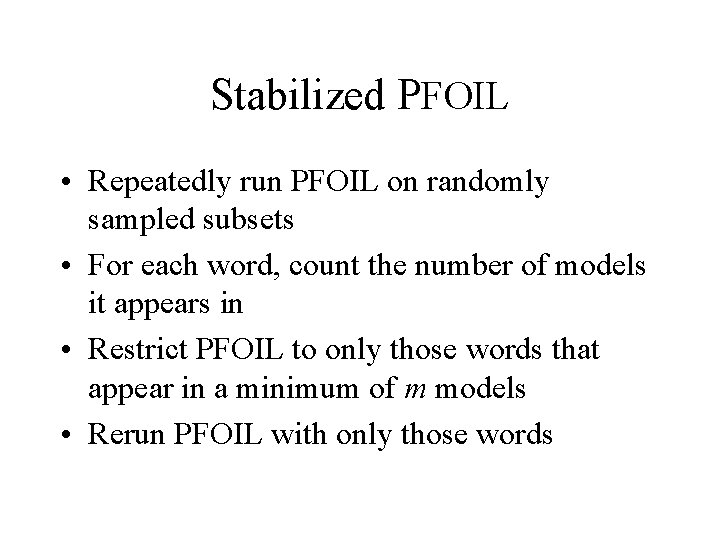
Stabilized PFOIL • Repeatedly run PFOIL on randomly sampled subsets • For each word, count the number of models it appears in • Restrict PFOIL to only those words that appear in a minimum of m models • Rerun PFOIL with only those words

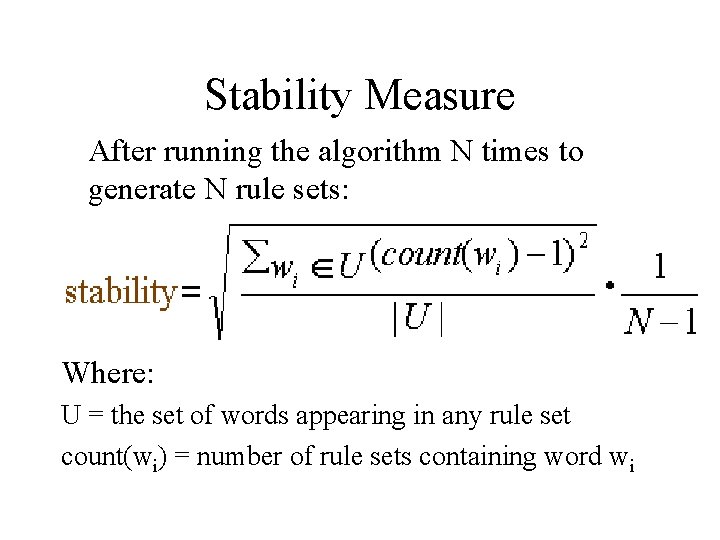
Stability Measure After running the algorithm N times to generate N rule sets: Where: U = the set of words appearing in any rule set count(wi) = number of rule sets containing word wi

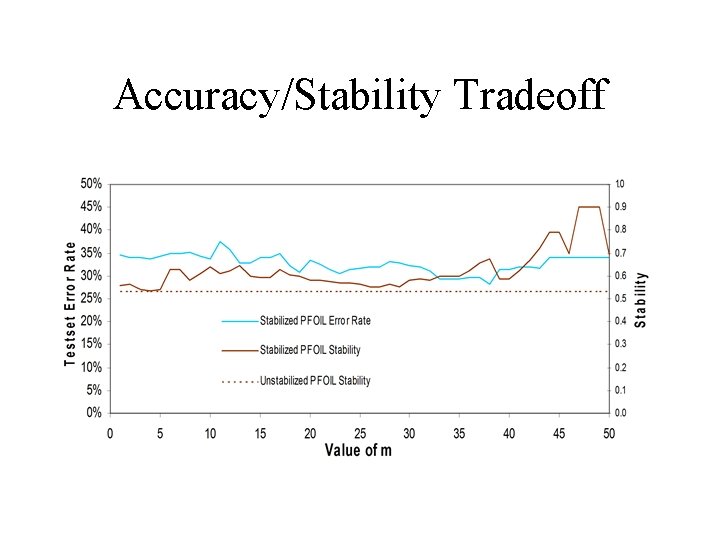
Accuracy/Stability Tradeoff

Discussion • Not very severe tradeoffs in Accuracy – vs. stability – vs. comprehensibility • PFOIL not as good at characterizing data – suggests not many dependencies – need for “softer” rules

Future Directions • M of N rules • Permutation Test • More Sources of Text Data

Take-Home Message • This is just a first step toward an aid for understanding expression data • Make expression models based on text in stead of DNA sequence.

Acknowledgements • This research was funded by the following grants: NLM 1 R 01 LM 07050 -01, NSF IRI-9502990, NIH 2 P 30 CA 14520 -29, and NIH 5 T 32 GM 08349.