International Symposium on the Recent Progress in Quantitative




- Slides: 30

International Symposium on the Recent Progress in Quantitative and Systems Biology , Dec 9 -11, The Chinese University of Hong Kong long-range allosteric effect in gene transcriptional regulation Ming Li Graduate School, CAS Zhong-can Ou-yang Institute of Theoretical Physics, CAS

Outline n Stressed state of DNA in vivo n Model of topologically constrained DNA and duplex separation n Long-range Allosteric Effect and Database investigation: a case study

Part I Stressed state of DNA in vivo

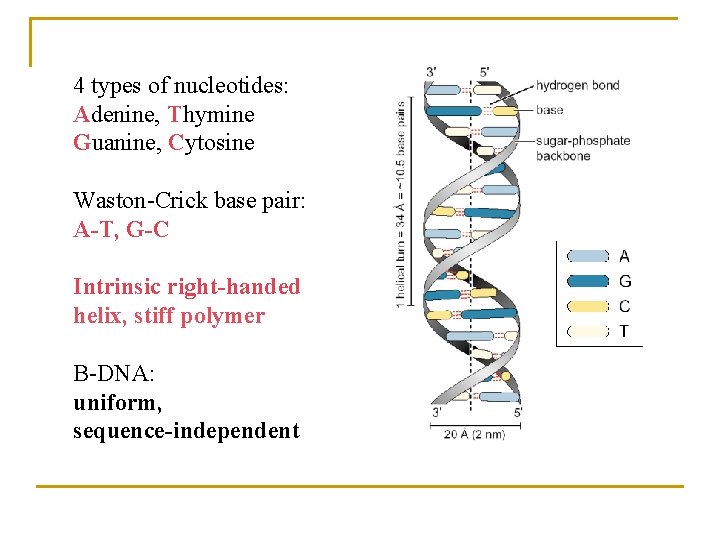
4 types of nucleotides: Adenine, Thymine Guanine, Cytosine Waston-Crick base pair: A-T, G-C Intrinsic right-handed helix, stiff polymer B-DNA: uniform, sequence-independent

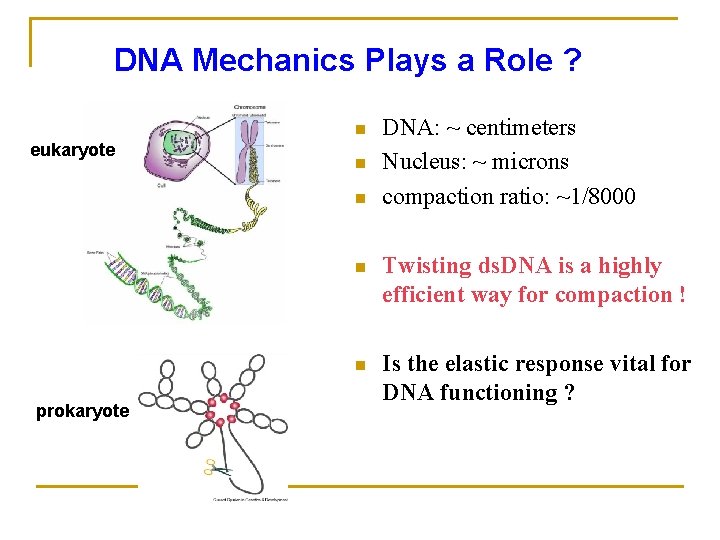
DNA Mechanics Plays a Role ? n eukaryote n n prokaryote DNA: ~ centimeters Nucleus: ~ microns compaction ratio: ~1/8000 n Twisting ds. DNA is a highly efficient way for compaction ! n Is the elastic response vital for DNA functioning ?

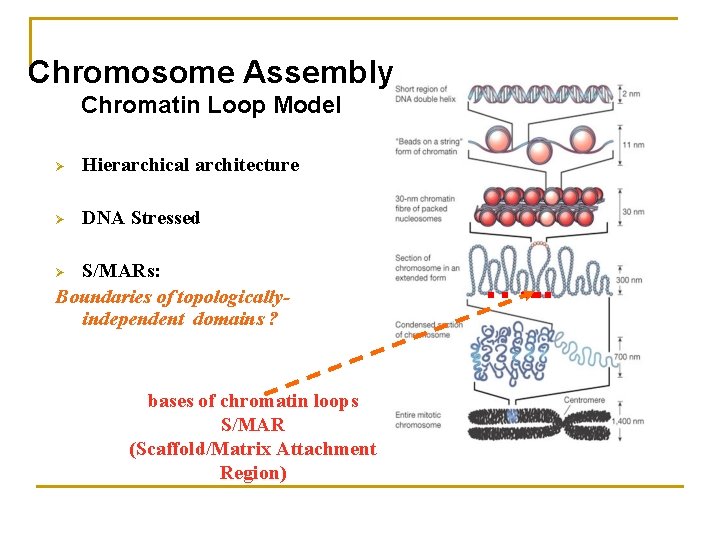
Chromosome Assembly Chromatin Loop Model Ø Hierarchical architecture Ø DNA Stressed S/MARs: Boundaries of topologicallyindependent domains ? Ø bases of chromatin loops S/MAR (Scaffold/Matrix Attachment Region)

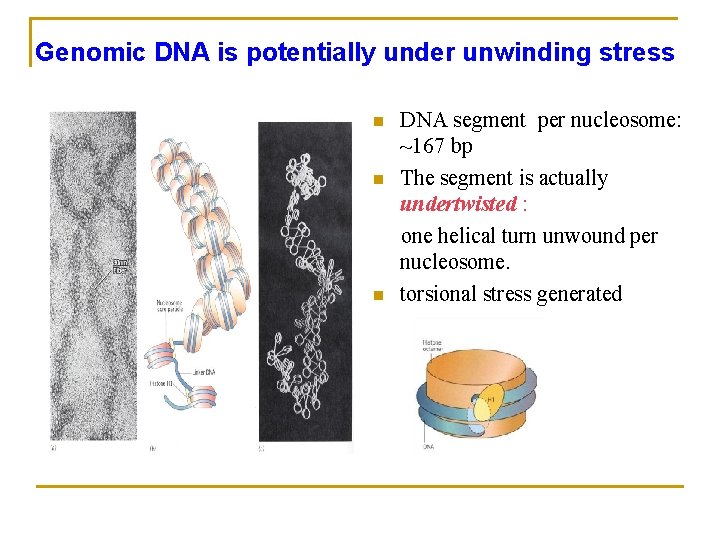
Genomic DNA is potentially under unwinding stress n n n DNA segment per nucleosome: ~167 bp The segment is actually undertwisted : one helical turn unwound per nucleosome. torsional stress generated

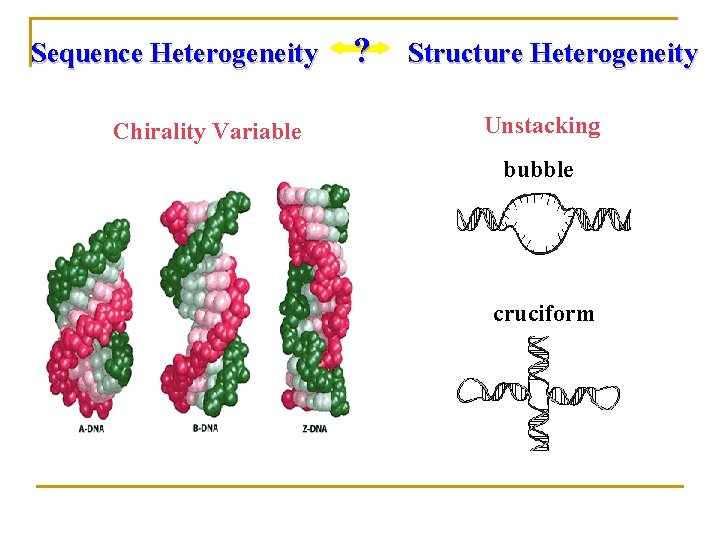
Sequence Heterogeneity Chirality Variable ? Structure Heterogeneity Unstacking bubble cruciform

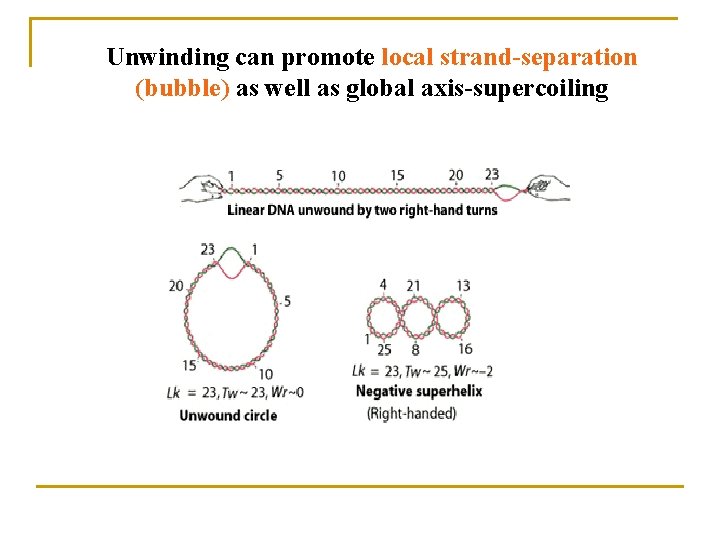
Unwinding can promote local strand-separation (bubble) as well as global axis-supercoiling

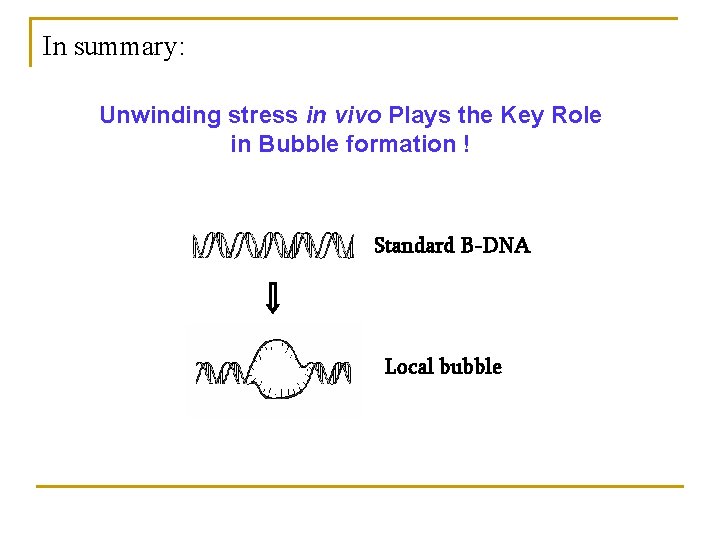
In summary: Unwinding stress in vivo Plays the Key Role in Bubble formation ! Standard B-DNA Local bubble

Part II Benham Model of topologically constrained DNA && sequence-dependent duplex separation

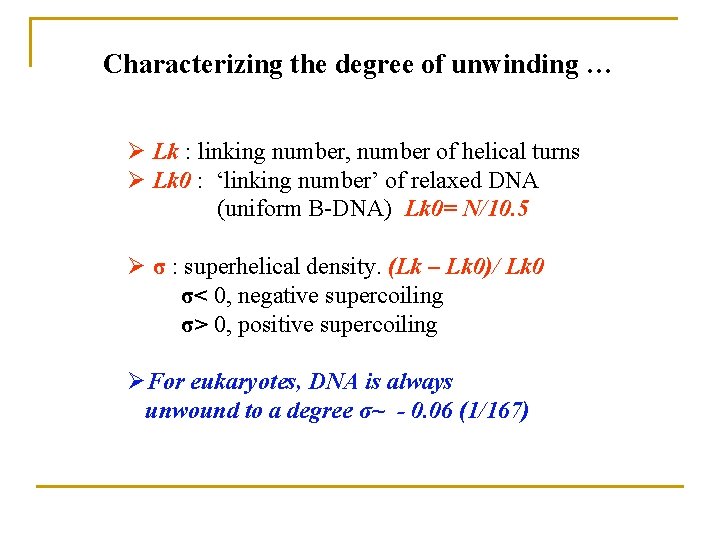
Characterizing the degree of unwinding … Ø Lk : linking number, number of helical turns Ø Lk 0 : ‘linking number’ of relaxed DNA (uniform B-DNA) Lk 0= N/10. 5 Ø σ : superhelical density. (Lk – Lk 0)/ Lk 0 σ< 0, negative supercoiling σ> 0, positive supercoiling ØFor eukaryotes, DNA is always unwound to a degree σ~ - 0. 06 (1/167)

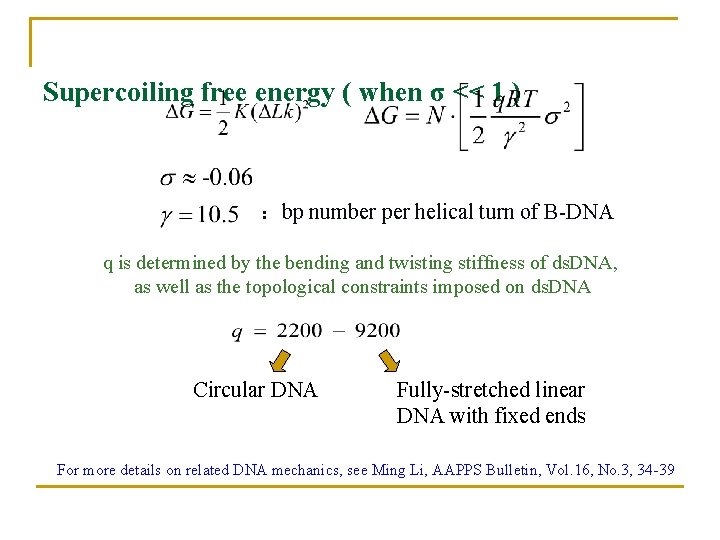
Supercoiling free energy ( when σ << 1 ) : bp number per helical turn of B-DNA q is determined by the bending and twisting stiffness of ds. DNA, as well as the topological constraints imposed on ds. DNA Circular DNA Fully-stretched linear DNA with fixed ends For more details on related DNA mechanics, see Ming Li, AAPPS Bulletin, Vol. 16, No. 3, 34 -39

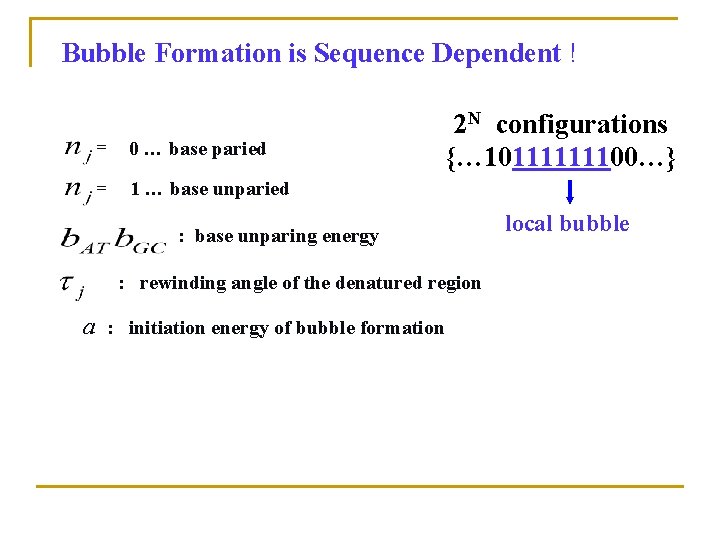
Bubble Formation is Sequence Dependent ! = 0 … base paried = 1 … base unparied 2 N configurations {… 10111111100…} : base unparing energy : rewinding angle of the denatured region a : initiation energy of bubble formation local bubble

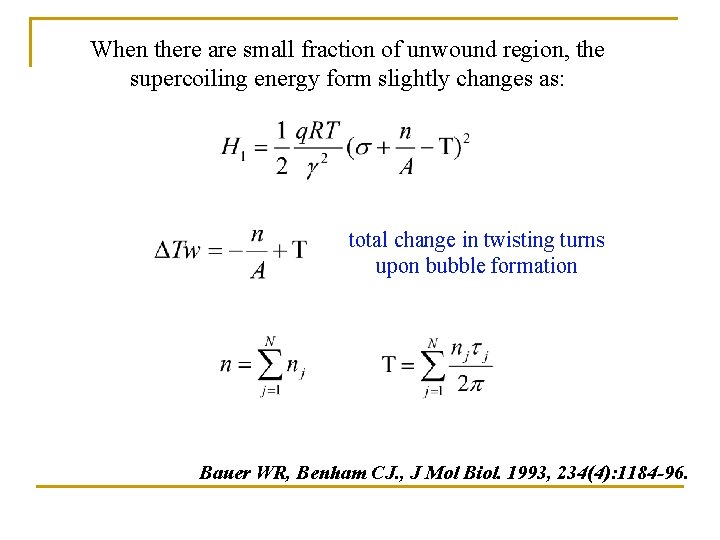
When there are small fraction of unwound region, the supercoiling energy form slightly changes as: total change in twisting turns upon bubble formation Bauer WR, Benham CJ. , J Mol Biol. 1993, 234(4): 1184 -96.

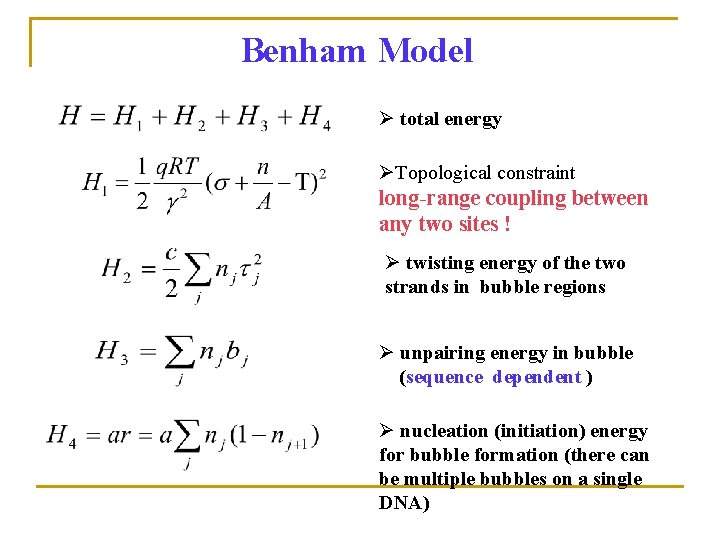
Benham Model Ø total energy ØTopological constraint long-range coupling between any two sites ! Ø twisting energy of the two strands in bubble regions Ø unpairing energy in bubble (sequence dependent ) Ø nucleation (initiation) energy for bubble formation (there can be multiple bubbles on a single DNA)

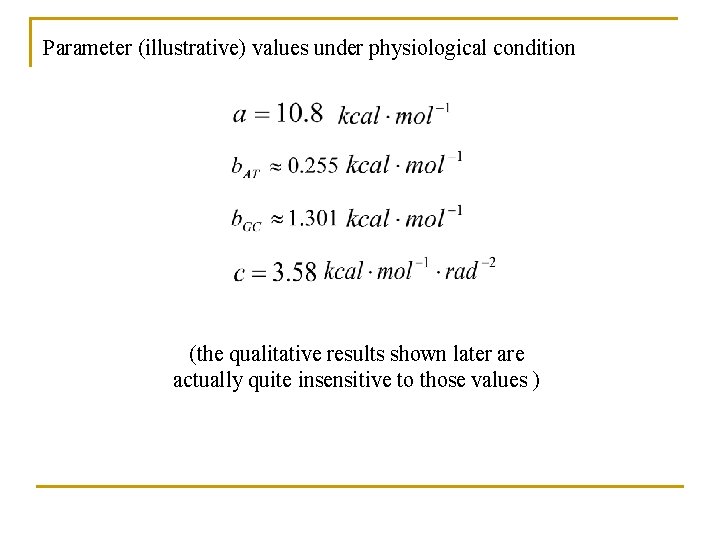
Parameter (illustrative) values under physiological condition (the qualitative results shown later are actually quite insensitive to those values )

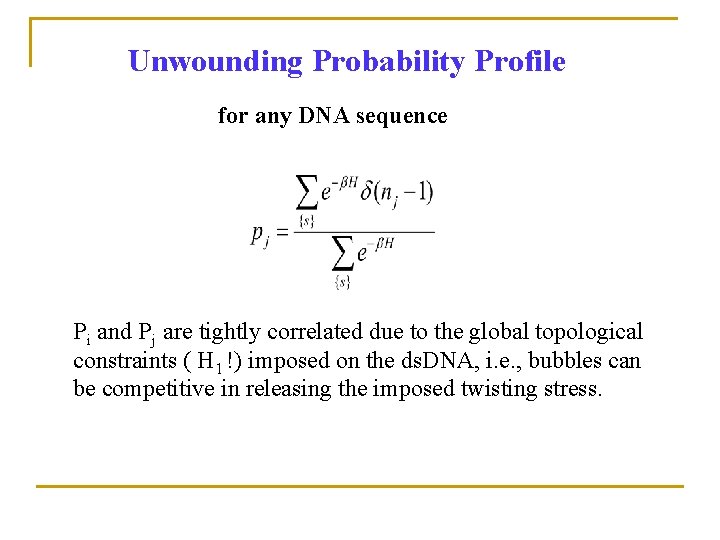
Unwounding Probability Profile for any DNA sequence Pi and Pj are tightly correlated due to the global topological constraints ( H 1 !) imposed on the ds. DNA, i. e. , bubbles can be competitive in releasing the imposed twisting stress.

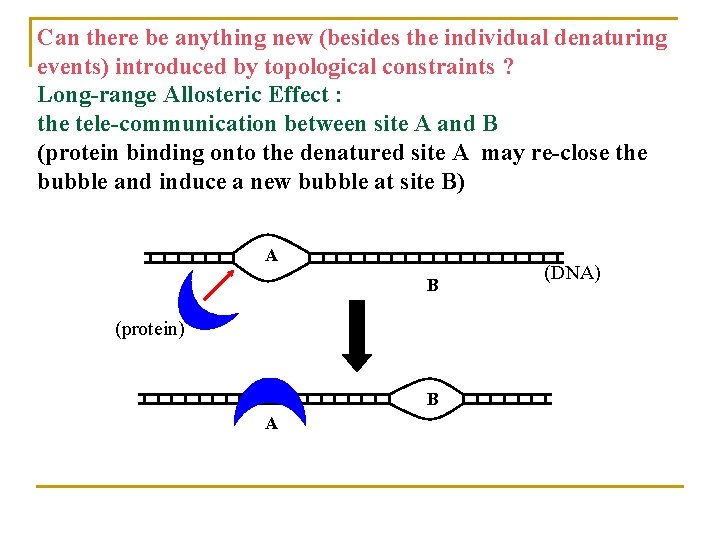
Can there be anything new (besides the individual denaturing events) introduced by topological constraints ? Long-range Allosteric Effect : the tele-communication between site A and B (protein binding onto the denatured site A may re-close the bubble and induce a new bubble at site B) A B (protein) B A (DNA)

Beginning with everything , ending with nothing ! It’s difficult to detect such a phenomenon in vivo by experiments, and it’s also almost impossible to directly ‘calculate’ such an effect quantitatively for real cases by taking account of every molecular detail (one can be drown in the details). Anyway, How can one do anything meaningful ? --- Bioinformatics to rescue ! Bioinformatics offers an alternative approach: exploring the biological data to find the statistically significant patterns which may cast some light on the understanding of the underlying molecular mechanism

Part III Long-range Allosteric Effect and Database investigation: a (bioinformatic) case study on SMAR function

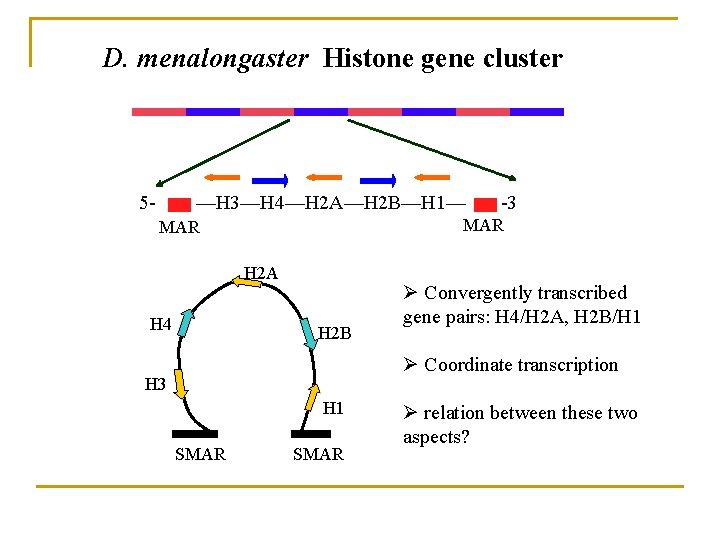
D. menalongaster Histone gene cluster 5 - —H 3—H 4—H 2 A—H 2 B—H 1— MAR H 2 A H 4 -3 H 2 B Ø Convergently transcribed gene pairs: H 4/H 2 A, H 2 B/H 1 Ø Coordinate transcription H 3 H 1 SMAR Ø relation between these two aspects?

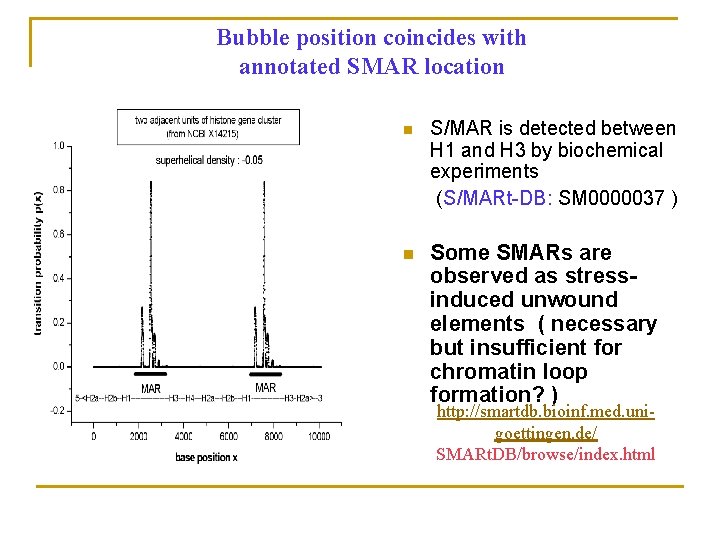
Bubble position coincides with annotated SMAR location n S/MAR is detected between H 1 and H 3 by biochemical experiments (S/MARt-DB: SM 0000037 ) n Some SMARs are observed as stressinduced unwound elements ( necessary but insufficient for chromatin loop formation? ) http: //smartdb. bioinf. med. unigoettingen. de/ SMARt. DB/browse/index. html

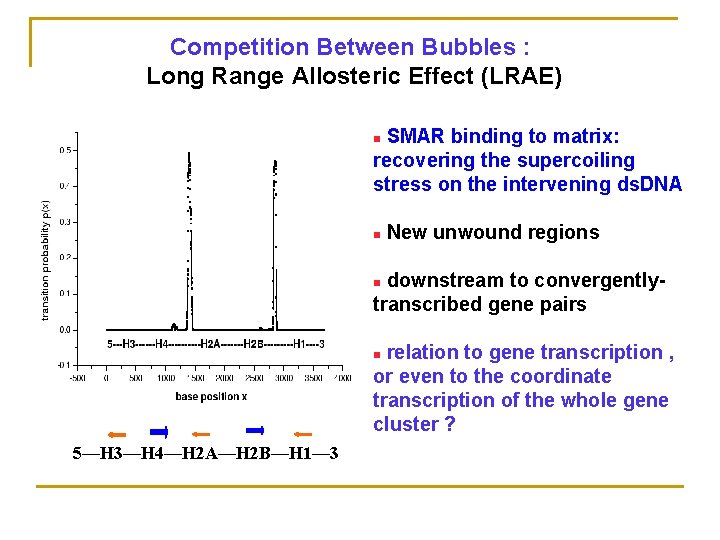
Competition Between Bubbles : Long Range Allosteric Effect (LRAE) SMAR binding to matrix: recovering the supercoiling stress on the intervening ds. DNA n n New unwound regions downstream to convergentlytranscribed gene pairs n relation to gene transcription , or even to the coordinate transcription of the whole gene cluster ? n 5—H 3—H 4—H 2 A—H 2 B—H 1— 3

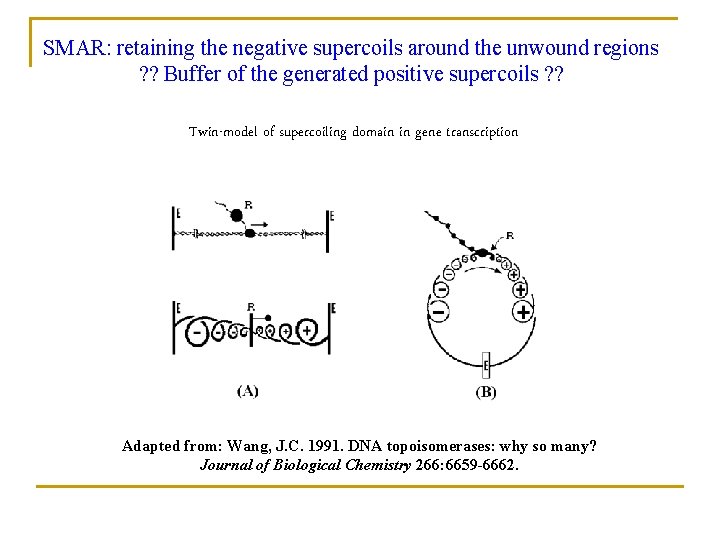
SMAR: retaining the negative supercoils around the unwound regions ? ? Buffer of the generated positive supercoils ? ? Twin-model of supercoiling domain in gene transcription Adapted from: Wang, J. C. 1991. DNA topoisomerases: why so many? Journal of Biological Chemistry 266: 6659 -6662.

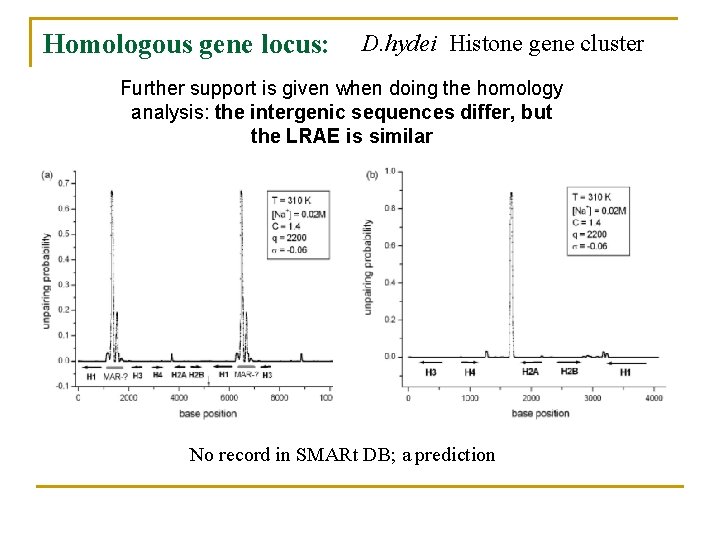
Homologous gene locus: D. hydei Histone gene cluster Further support is given when doing the homology analysis: the intergenic sequences differ, but the LRAE is similar No record in SMARt DB; a prediction

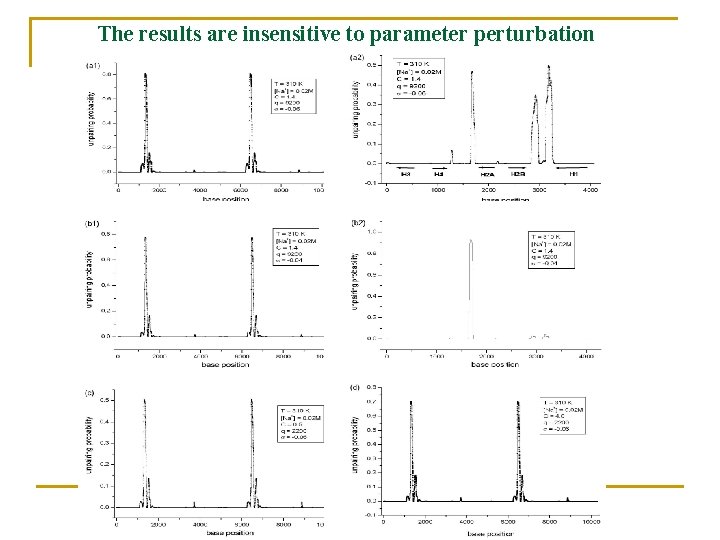
The results are insensitive to parameter perturbation

More examples supporting the existence of LRAE : Sorghum v. s. Rice : Sh 2/A 1 (homologous locus) SM 0000063 SM 0000064 SM 0000065 SM 0000066 SM 0000067 SM 0000068 SM 0000069 SM 0000070 SM 0000071 Sorghum v. s. Maize : Adh 1 (homologous locus) SM 0000032 SM 00000170 hs. IFNA SM 0000011 SM 0000012 IGF 2 SM 0000023 SM 0000028 SM 0000029 IFNA 2 SM 0000074 SM 0000075 csp. B SM 0000080 SM 0000081 S/MARt-DB is still under construction. Meaningful statistics should be given when there are enough records available.

Summary n It’s possible to reveal LRAE by sequence analysis combined with mechanics investigation in a bioinformatic way n LRAE is hopefully an effective regulatory mechanism in gene transcription (e. g. SMAR can function via LRAE)

Thanks For Your Attention !