Integrating Free Surfer and FSL FEAT 1 Outline







- Slides: 36

Integrating Free. Surfer and FSL FEAT 1

Outline • Registering FEAT Free. Surfer Anatomical • Automatic (FLIRT) • Manual (tkregister 2) • Surface-based Group f. MRI Analysis • Individual Analysis • Viewing FEAT output on Anatomical • Sampling FEAT output on the surface • Mapping Free. Surfer Segmentations to FEAT 2

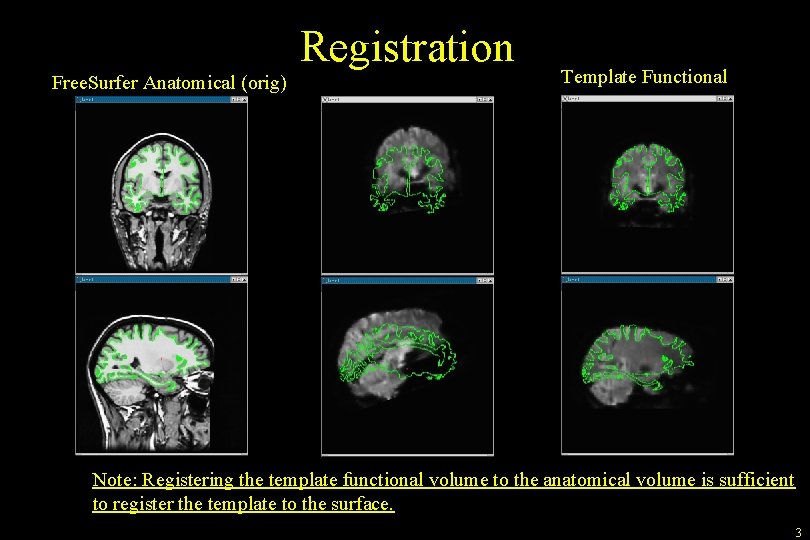
Registration Free. Surfer Anatomical (orig) Template Functional Note: Registering the template functional volume to the anatomical volume is sufficient to register the template to the surface. 3

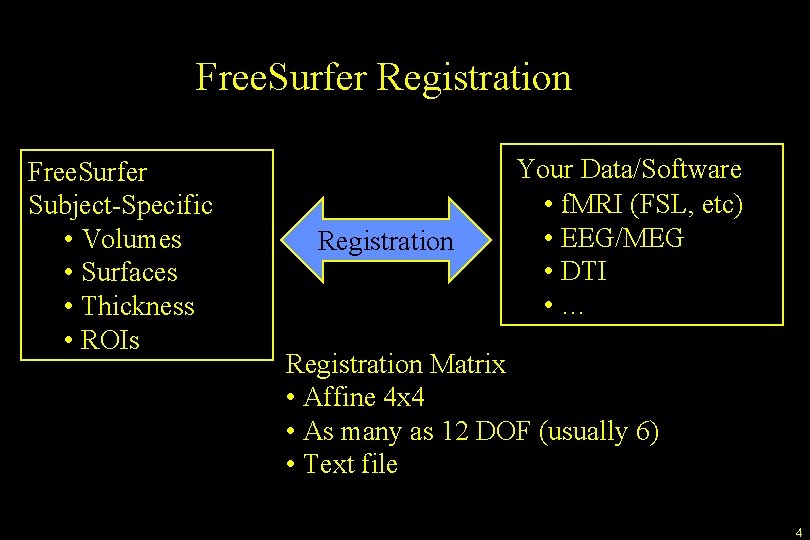
Free. Surfer Registration Free. Surfer Subject-Specific • Volumes • Surfaces • Thickness • ROIs Registration Your Data/Software • f. MRI (FSL, etc) • EEG/MEG • DTI • … Registration Matrix • Affine 4 x 4 • As many as 12 DOF (usually 6) • Text file 4

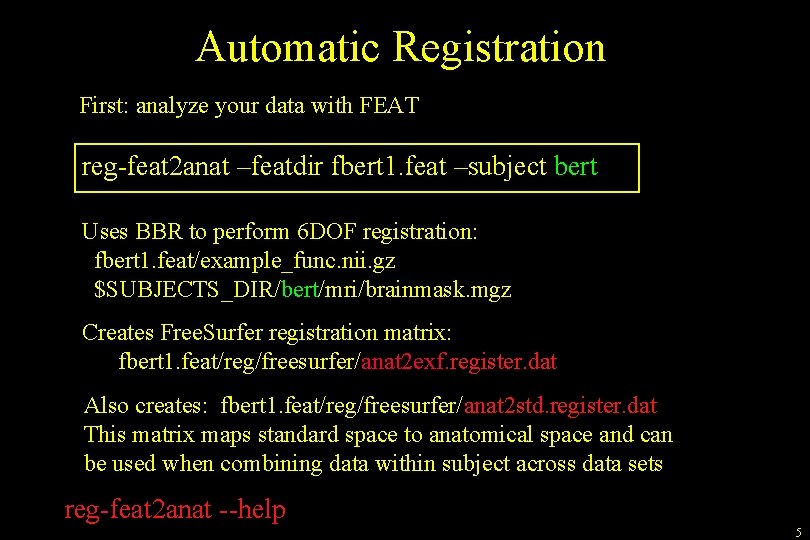
Automatic Registration First: analyze your data with FEAT reg-feat 2 anat –featdir fbert 1. feat –subject bert Uses BBR to perform 6 DOF registration: fbert 1. feat/example_func. nii. gz $SUBJECTS_DIR/bert/mri/brainmask. mgz Creates Free. Surfer registration matrix: fbert 1. feat/reg/freesurfer/anat 2 exf. register. dat Also creates: fbert 1. feat/reg/freesurfer/anat 2 std. register. dat This matrix maps standard space to anatomical space and can be used when combining data within subject across data sets reg-feat 2 anat --help 5

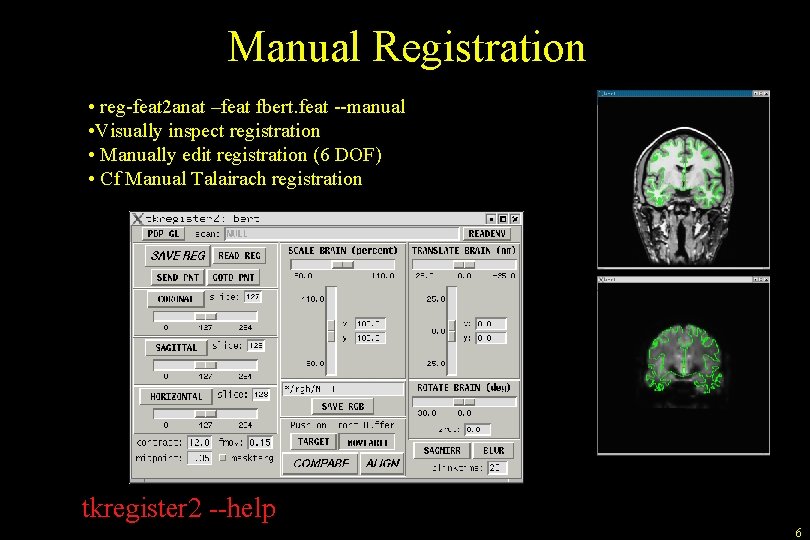
Manual Registration • reg-feat 2 anat –feat fbert. feat --manual • Visually inspect registration • Manually edit registration (6 DOF) • Cf Manual Talairach registration tkregister 2 --help 6

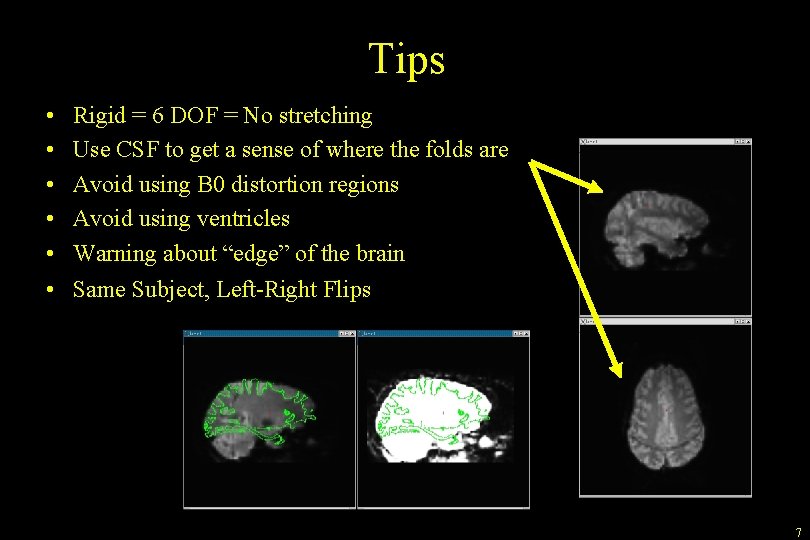
Tips • • • Rigid = 6 DOF = No stretching Use CSF to get a sense of where the folds are Avoid using B 0 distortion regions Avoid using ventricles Warning about “edge” of the brain Same Subject, Left-Right Flips 7

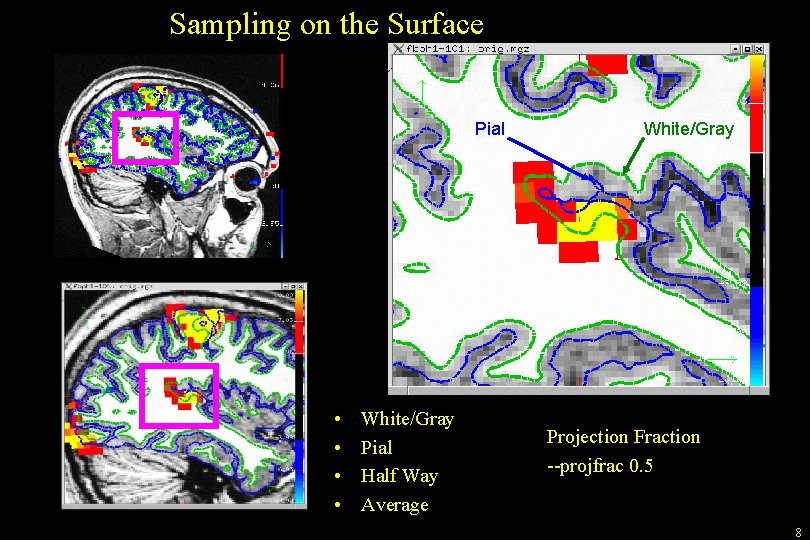
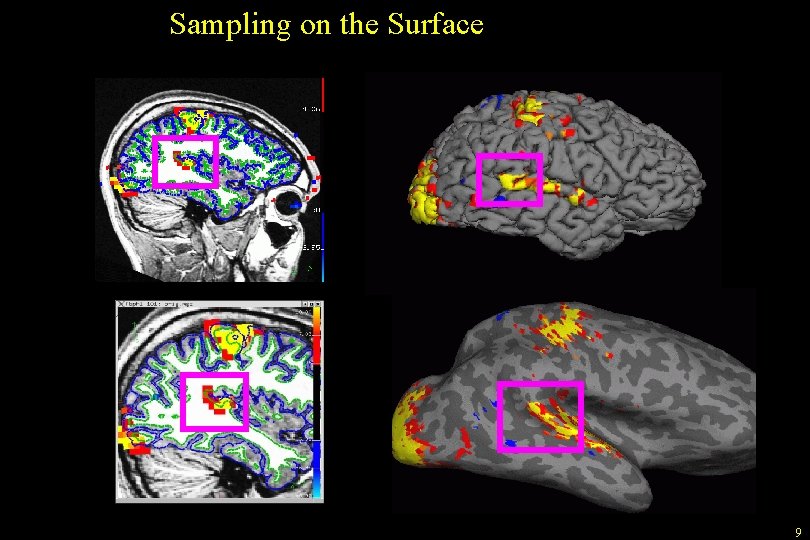
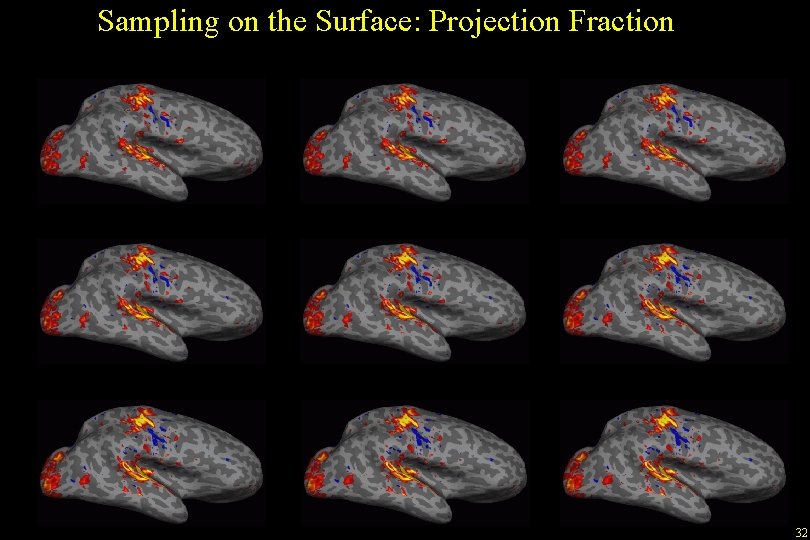
Sampling on the Surface Pial • • White/Gray Pial Half Way Average White/Gray Projection Fraction --projfrac 0. 5 8

Sampling on the Surface 9

Surface-based f. MRI Group Analysis 10

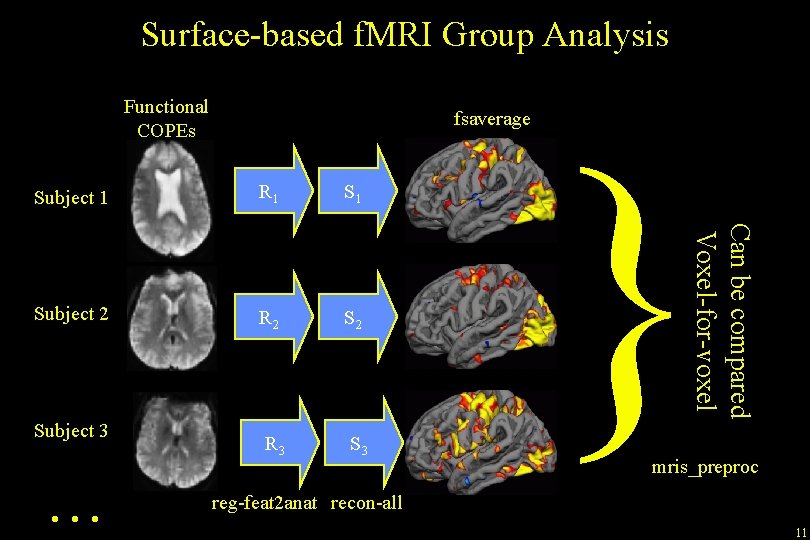
Surface-based f. MRI Group Analysis Functional COPEs fsaverage R 1 Subject 2 R 2 S 2 R 3 Subject 3 … } Can be compared Voxel-for-voxel Subject 1 mris_preproc reg-feat 2 anat recon-all 11

Surface-based f. MRI Group Analysis (One Run) • Single Run • Analyze with FEAT, No smoothing • COPEs are in native functional space Subject 1 run 1. feat stats cope 1. nii 12

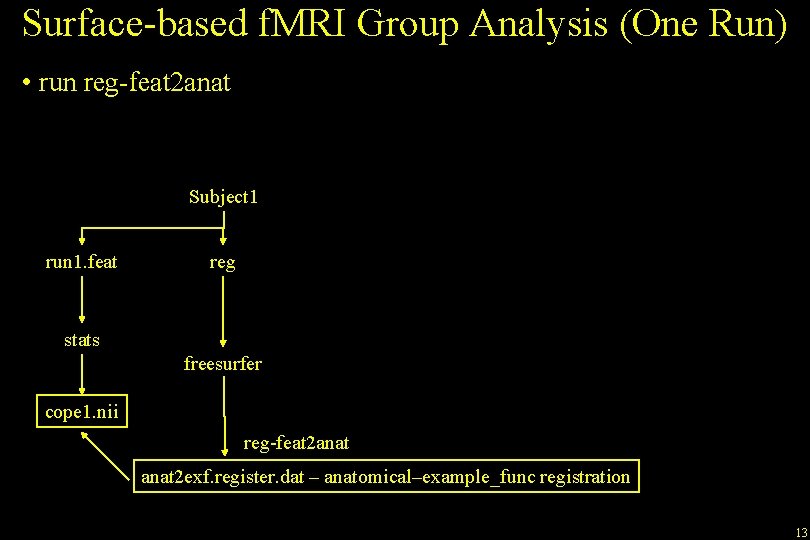
Surface-based f. MRI Group Analysis (One Run) • run reg-feat 2 anat Subject 1 run 1. feat reg stats freesurfer cope 1. nii reg-feat 2 anat 2 exf. register. dat – anatomical–example_func registration 13

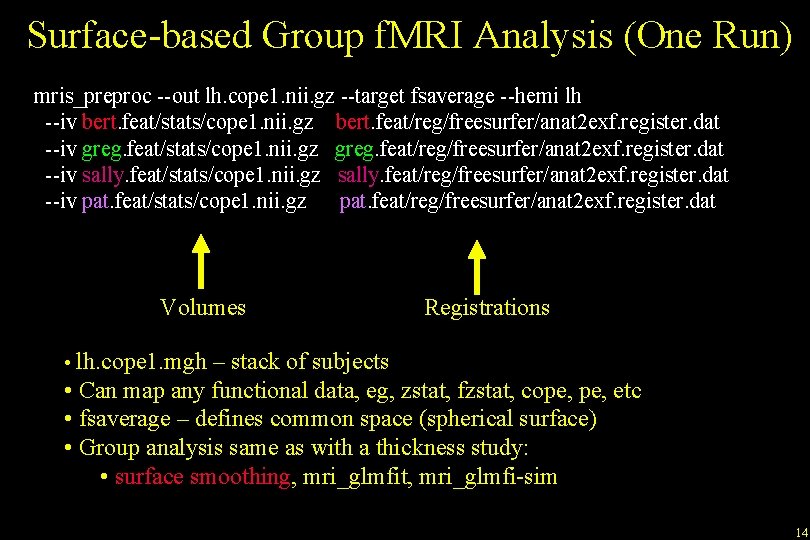
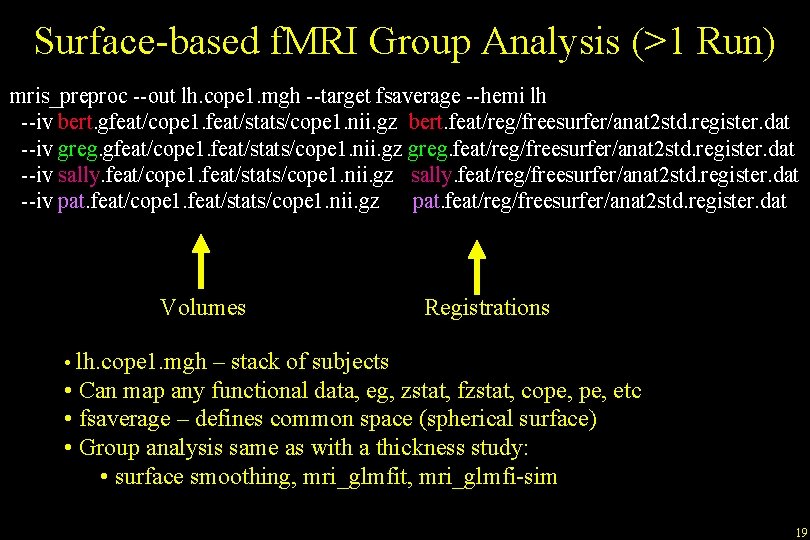
Surface-based Group f. MRI Analysis (One Run) mris_preproc --out lh. cope 1. nii. gz --target fsaverage --hemi lh --iv bert. feat/stats/cope 1. nii. gz bert. feat/reg/freesurfer/anat 2 exf. register. dat --iv greg. feat/stats/cope 1. nii. gz greg. feat/reg/freesurfer/anat 2 exf. register. dat --iv sally. feat/stats/cope 1. nii. gz sally. feat/reg/freesurfer/anat 2 exf. register. dat --iv pat. feat/stats/cope 1. nii. gz pat. feat/reg/freesurfer/anat 2 exf. register. dat Volumes Registrations • lh. cope 1. mgh – stack of subjects • Can map any functional data, eg, zstat, fzstat, cope, etc • fsaverage – defines common space (spherical surface) • Group analysis same as with a thickness study: • surface smoothing, mri_glmfit, mri_glmfi-sim 14

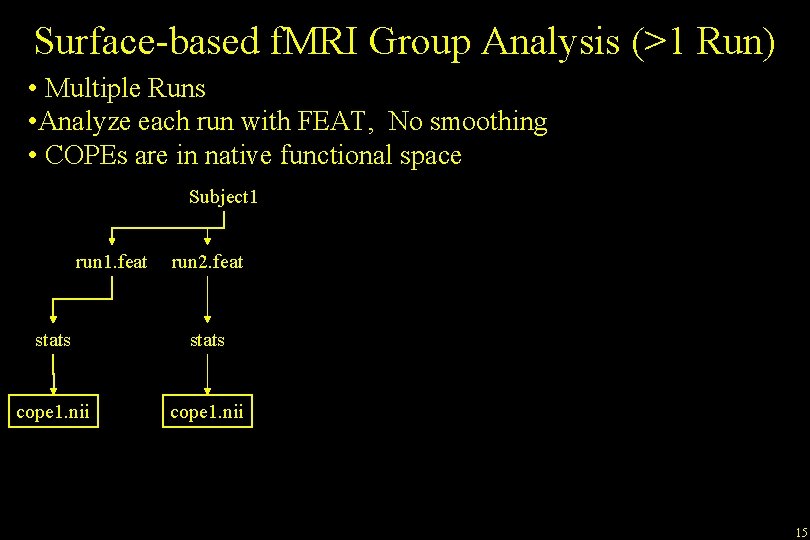
Surface-based f. MRI Group Analysis (>1 Run) • Multiple Runs • Analyze each run with FEAT, No smoothing • COPEs are in native functional space Subject 1 run 1. feat run 2. feat stats cope 1. nii 15

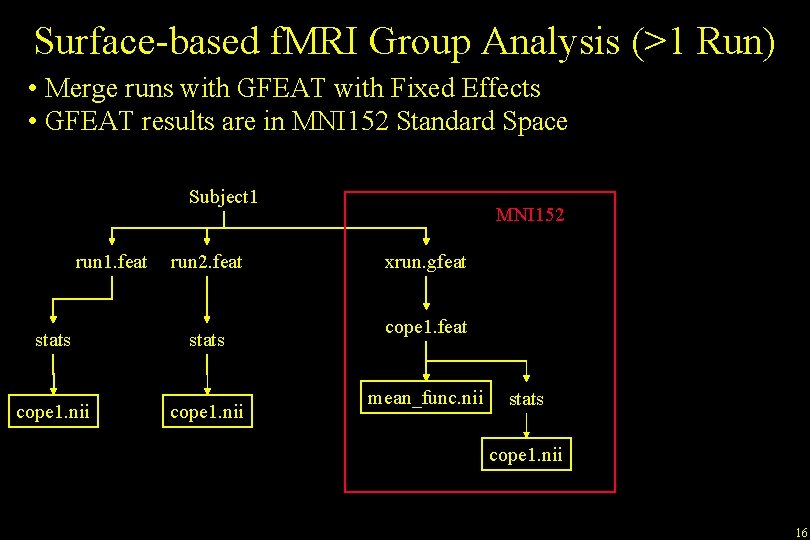
Surface-based f. MRI Group Analysis (>1 Run) • Merge runs with GFEAT with Fixed Effects • GFEAT results are in MNI 152 Standard Space Subject 1 run 1. feat stats cope 1. nii run 2. feat stats cope 1. nii MNI 152 xrun. gfeat cope 1. feat mean_func. nii stats cope 1. nii 16

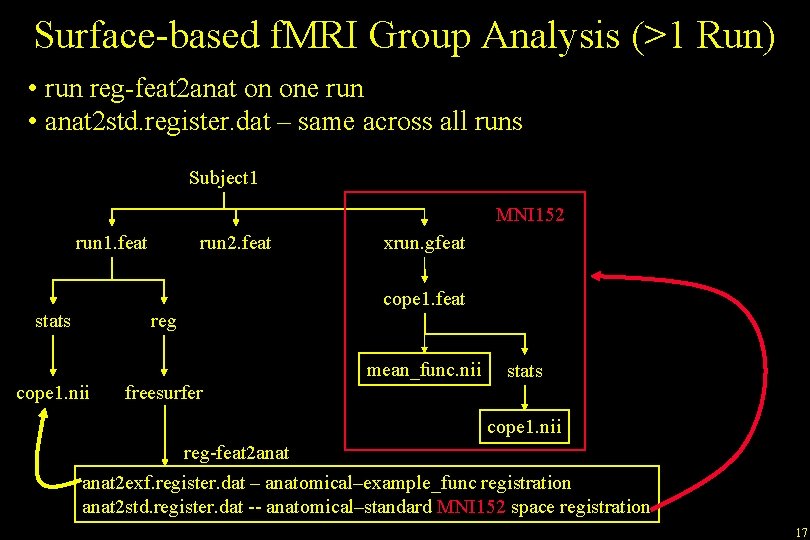
Surface-based f. MRI Group Analysis (>1 Run) • run reg-feat 2 anat on one run • anat 2 std. register. dat – same across all runs Subject 1 MNI 152 run 1. feat stats run 2. feat reg xrun. gfeat cope 1. feat mean_func. nii cope 1. nii freesurfer stats cope 1. nii reg-feat 2 anat 2 exf. register. dat – anatomical–example_func registration anat 2 std. register. dat -- anatomical–standard MNI 152 space registration 17

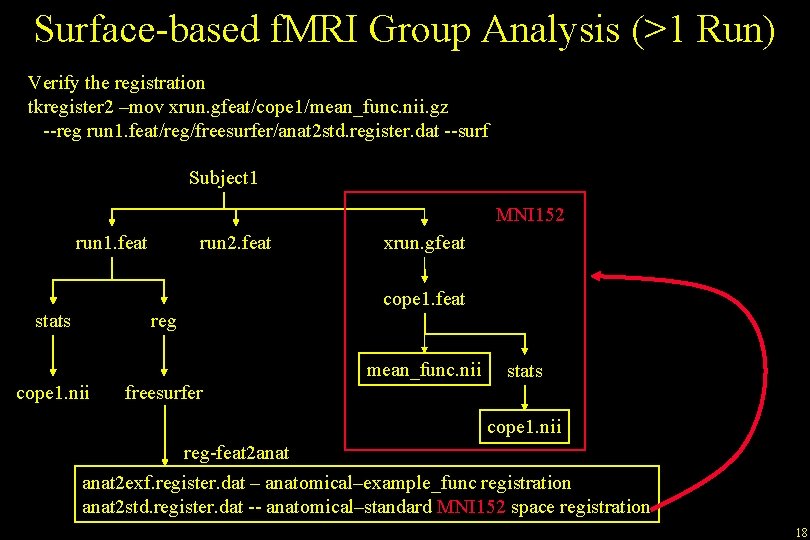
Surface-based f. MRI Group Analysis (>1 Run) Verify the registration tkregister 2 –mov xrun. gfeat/cope 1/mean_func. nii. gz --reg run 1. feat/reg/freesurfer/anat 2 std. register. dat --surf Subject 1 MNI 152 run 1. feat stats run 2. feat reg xrun. gfeat cope 1. feat mean_func. nii cope 1. nii freesurfer stats cope 1. nii reg-feat 2 anat 2 exf. register. dat – anatomical–example_func registration anat 2 std. register. dat -- anatomical–standard MNI 152 space registration 18

Surface-based f. MRI Group Analysis (>1 Run) mris_preproc --out lh. cope 1. mgh --target fsaverage --hemi lh --iv bert. gfeat/cope 1. feat/stats/cope 1. nii. gz bert. feat/reg/freesurfer/anat 2 std. register. dat --iv greg. gfeat/cope 1. feat/stats/cope 1. nii. gz greg. feat/reg/freesurfer/anat 2 std. register. dat --iv sally. feat/cope 1. feat/stats/cope 1. nii. gz sally. feat/reg/freesurfer/anat 2 std. register. dat --iv pat. feat/cope 1. feat/stats/cope 1. nii. gz pat. feat/reg/freesurfer/anat 2 std. register. dat Volumes Registrations • lh. cope 1. mgh – stack of subjects • Can map any functional data, eg, zstat, fzstat, cope, etc • fsaverage – defines common space (spherical surface) • Group analysis same as with a thickness study: • surface smoothing, mri_glmfit, mri_glmfi-sim 19

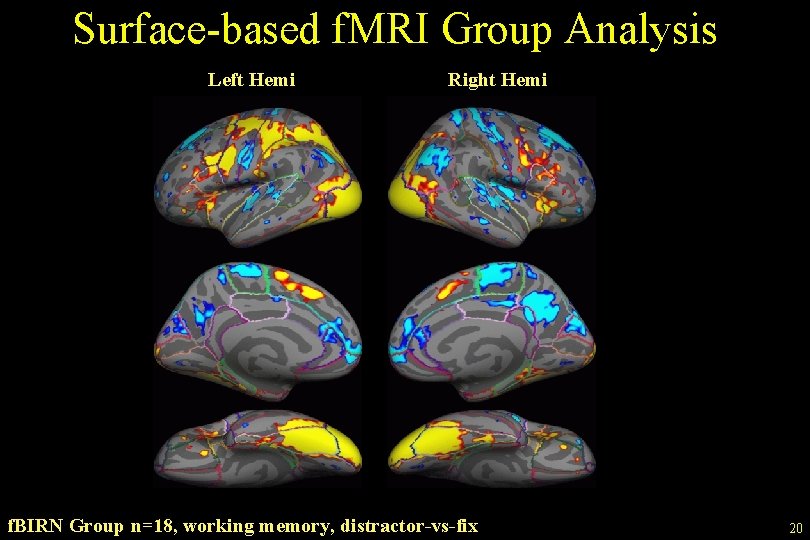
Surface-based f. MRI Group Analysis Left Hemi Right Hemi f. BIRN Group n=18, working memory, distractor-vs-fix 20

Individual Subject Integration Applying Free. Surfer Tools to FSL f. MRI Analysis 21

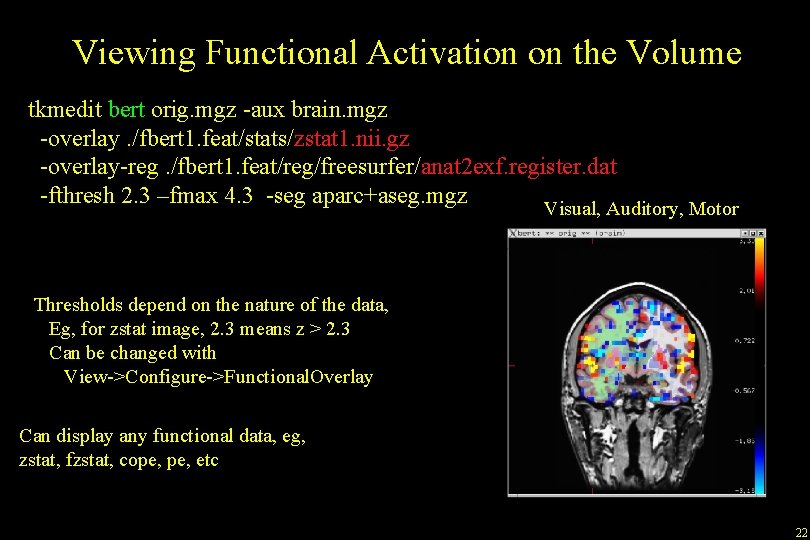
Viewing Functional Activation on the Volume tkmedit bert orig. mgz -aux brain. mgz -overlay. /fbert 1. feat/stats/zstat 1. nii. gz -overlay-reg. /fbert 1. feat/reg/freesurfer/anat 2 exf. register. dat -fthresh 2. 3 –fmax 4. 3 -seg aparc+aseg. mgz Visual, Auditory, Motor Thresholds depend on the nature of the data, Eg, for zstat image, 2. 3 means z > 2. 3 Can be changed with View->Configure->Functional. Overlay Can display any functional data, eg, zstat, fzstat, cope, etc 22

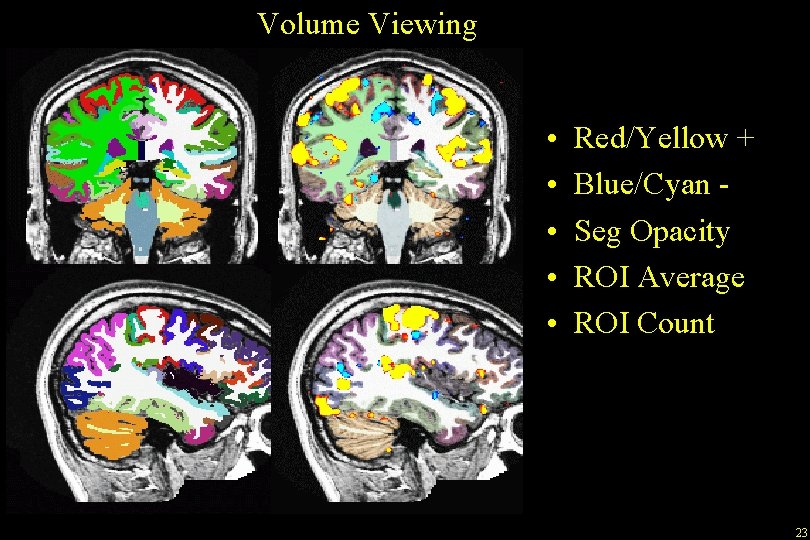
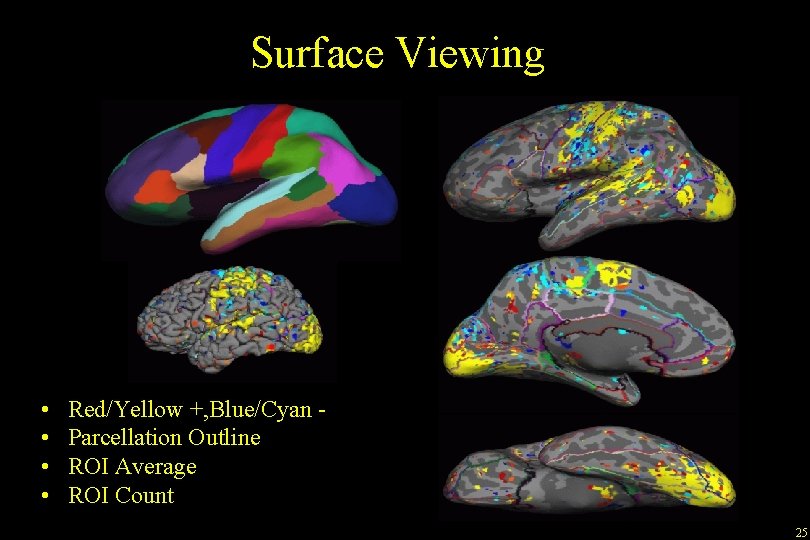
Volume Viewing • • • Red/Yellow + Blue/Cyan Seg Opacity ROI Average ROI Count 23

Viewing FEAT Stats on the Surface tksurfer bert rh inflated -overlay. /fbert 1. feat/stats/zstat 1. nii. gz -overlay-reg. /fbert 1. feat/reg/freesurfer/anat 2 exf. register. dat -fthresh 2. 3 -fmid 3. 3 -fslope 1 Visual, Auditory, Motor Can display any functional data, eg, zstat, fzstat, cope, etc 24

Surface Viewing • • Red/Yellow +, Blue/Cyan Parcellation Outline ROI Average ROI Count 25

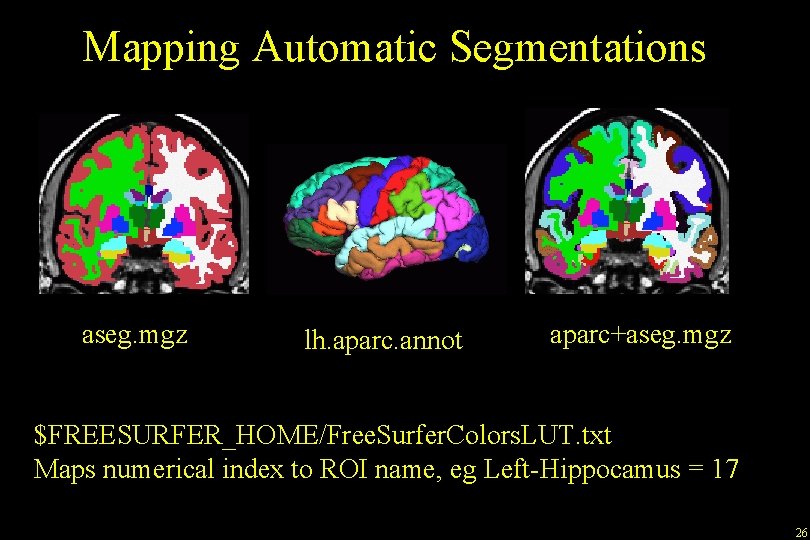
Mapping Automatic Segmentations aseg. mgz lh. aparc. annot aparc+aseg. mgz $FREESURFER_HOME/Free. Surfer. Colors. LUT. txt Maps numerical index to ROI name, eg Left-Hippocamus = 17 26

Mapping Automatic Segmentations aseg 2 feat --feat fbert. feat –aseg aparc+aseg In the functional FOV, creates: fbert 1. feat/reg/freesurfer/aseg+aparc. nii. gz Create Text Summary Table (nvox, mean cope, std cope, etc) mri_segstats --seg fbert 1. feat/reg/freesurfer/aparc+aseg. nii. gz --nonempty --ctab-default --in fbert. feat/stats/cope 1. nii. gz --sum fbert 1. sum. txt Can summarize any functional data, eg, zstat, fzstat, cope, etc 27

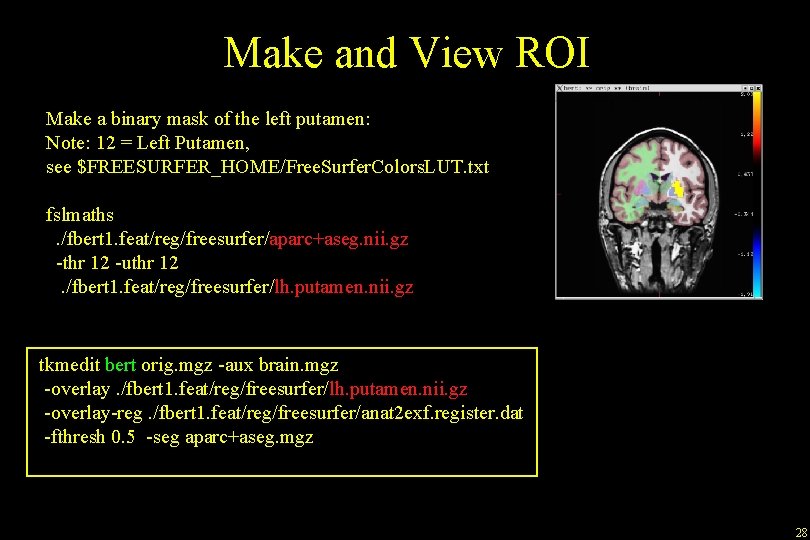
Make and View ROI Make a binary mask of the left putamen: Note: 12 = Left Putamen, see $FREESURFER_HOME/Free. Surfer. Colors. LUT. txt fslmaths. /fbert 1. feat/reg/freesurfer/aparc+aseg. nii. gz -thr 12 -uthr 12. /fbert 1. feat/reg/freesurfer/lh. putamen. nii. gz tkmedit bert orig. mgz -aux brain. mgz -overlay. /fbert 1. feat/reg/freesurfer/lh. putamen. nii. gz -overlay-reg. /fbert 1. feat/reg/freesurfer/anat 2 exf. register. dat -fthresh 0. 5 -seg aparc+aseg. mgz 28

Practical Data • • • Sensory-motor study Blocked Design (15 sec OFF, 15 sec ON) One subject – “bert” Two runs (TR=3, 85 time points) FEAT has been run on both runs (FWHM=5) Combined with GFEAT – FFx – One-Sample Group Mean (OSGM) • Actually, all analysis steps already performed! 29

Practical • • Automatic registration (<5 min) Manual registration View FEAT results on subject’s anatomy (aparc+aseg) View FEAT results with tksurfer Map aparc+aseg to Functional Space Verify GFEAT registration View GFEAT results in volume and on surface “Higher-Level” analysis with mri_glmfit – Cross-Run – Fixed-Effects (FFX), One-Sample Group Mean (osgm) 30

End of Presentation 31

Sampling on the Surface: Projection Fraction -0. 1 0. 0 (white) +0. 1 +0. 3 +0. 5 +0. 7 +0. 9 +1. 0 (pial) +1. 1 32

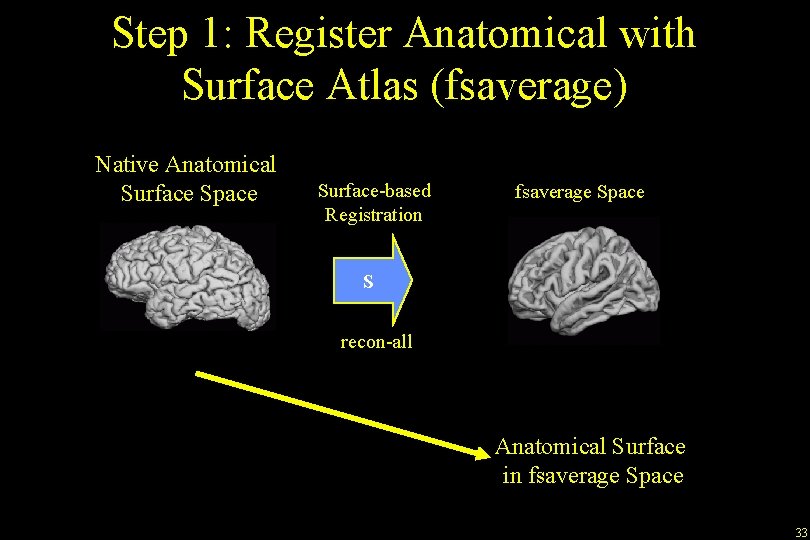
Step 1: Register Anatomical with Surface Atlas (fsaverage) Native Anatomical Surface Space Surface-based Registration fsaverage Space S recon-all Anatomical Surface in fsaverage Space 33

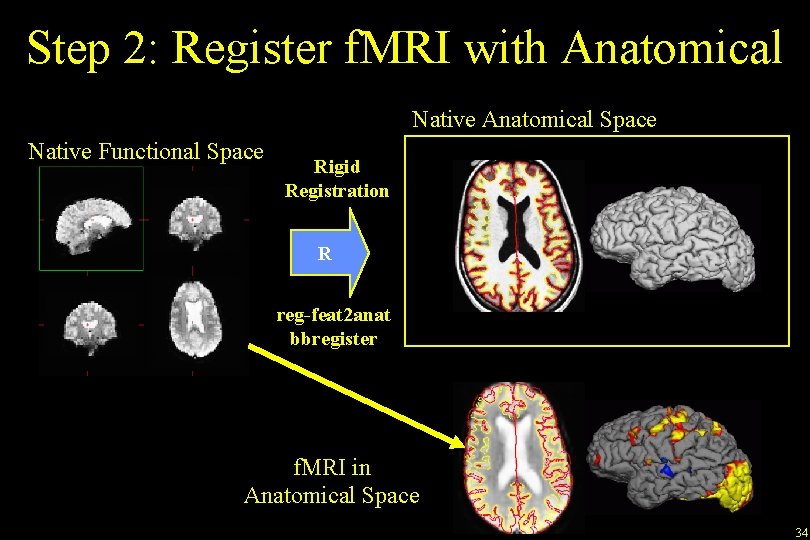
Step 2: Register f. MRI with Anatomical Native Anatomical Space Native Functional Space Rigid Registration R reg-feat 2 anat bbregister f. MRI in Anatomical Space 34

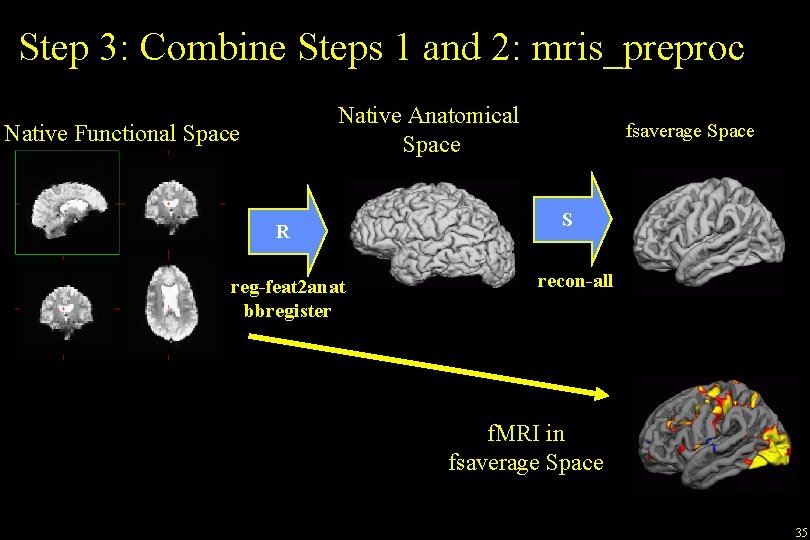
Step 3: Combine Steps 1 and 2: mris_preproc Native Anatomical Space Native Functional Space R reg-feat 2 anat bbregister fsaverage Space S recon-all f. MRI in fsaverage Space 35

Within-Subject, Cross-Run Analysis • Analyze each run with FEAT (data. X. feat) • Combine runs with GFEAT (standard space) • mean_func. nii. gz – avg of example_func in std space • Register each run (not. gfeat) with reg-feat 2 anat. • data. X. feat/reg/freesurfer/anat 2 std. register. dat • All runs (X) should be very close Verify the registration tkregister 2 –mov data. gfeat/mean_func. nii. gz --reg anat 2 std. register. dat --surf Use anat 2 std. register. dat the way you would anat 2 exf. register. dat 36
 Free surfer
Free surfer Free surfer meaning
Free surfer meaning Fsl blaze
Fsl blaze Fsl vs spm
Fsl vs spm Fsl roi
Fsl roi Fsl fmri
Fsl fmri Tdsb fsl
Tdsb fsl A surfer hangs ten and accelerates
A surfer hangs ten and accelerates Surfer kriging
Surfer kriging Surfer
Surfer Surfer
Surfer Surfer
Surfer Surfer
Surfer Mris_preproc
Mris_preproc Surfer
Surfer Surfer
Surfer Random surfer model
Random surfer model Surfer's myelopathy
Surfer's myelopathy Interpersonal skills in soul surfer
Interpersonal skills in soul surfer Fermi surfer
Fermi surfer Libtetrabz
Libtetrabz Quote sandwich examples
Quote sandwich examples Integrating classification and association rule mining
Integrating classification and association rule mining Key internal forces
Key internal forces Non exact ode calculator
Non exact ode calculator Integrating science and social studies
Integrating science and social studies Integrating strategy and culture in internal assessment
Integrating strategy and culture in internal assessment Integrating qualitative and quantitative methods
Integrating qualitative and quantitative methods Integrating communications assessment and tactics
Integrating communications assessment and tactics Integrating sel and pbis
Integrating sel and pbis Integrating public health and primary care
Integrating public health and primary care Helmholtz free energy
Helmholtz free energy Summary of story of an hour
Summary of story of an hour Free hearts free foreheads you and i are
Free hearts free foreheads you and i are Integrating quotes into an essay
Integrating quotes into an essay Integrating quotes mla
Integrating quotes mla Longitudinal fissure
Longitudinal fissure