Integrated transcriptional profiling and linkage analysis for mapping





- Slides: 37

Integrated transcriptional profiling and linkage analysis for mapping disease genes and regulatory gene networks analysis Enrico Petretto Research Fellow in Genomic Medicine Imperial College Faculty of Medicine enrico. petretto@imperial. ac. uk

Outline • Introduction: the biological framework – Expression QTL mapping using animal models – e. QTL analysis in multiple tissues • Integrating genome-wide e. QTL data to identify gene association networks – Data mining of e. QTLs – Graphical Gaussian models (GGMs) – Example of identification of disregulated pathway – Master transcriptional regulator

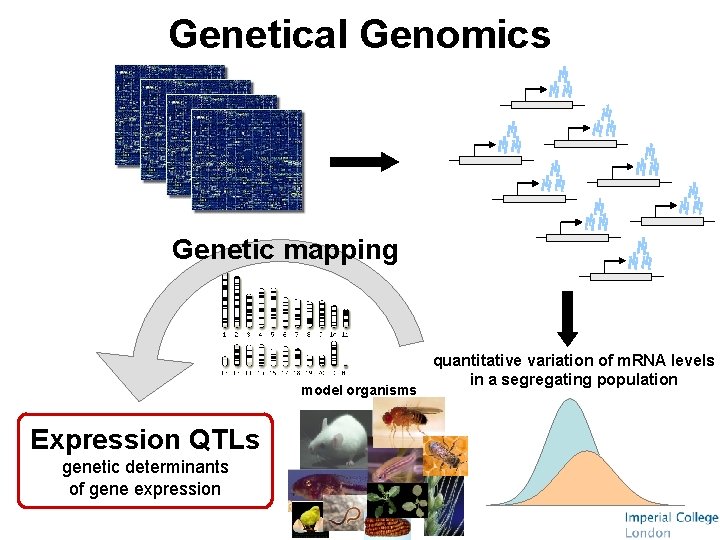
Genetical Genomics Genetic mapping model organisms Expression QTLs genetic determinants of gene expression quantitative variation of m. RNA levels in a segregating population

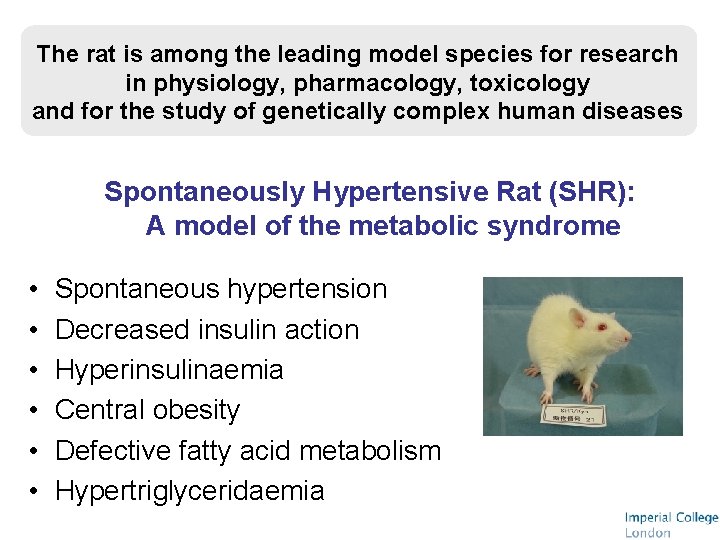
The rat is among the leading model species for research in physiology, pharmacology, toxicology and for the study of genetically complex human diseases Spontaneously Hypertensive Rat (SHR): A model of the metabolic syndrome • • • Spontaneous hypertension Decreased insulin action Hyperinsulinaemia Central obesity Defective fatty acid metabolism Hypertriglyceridaemia

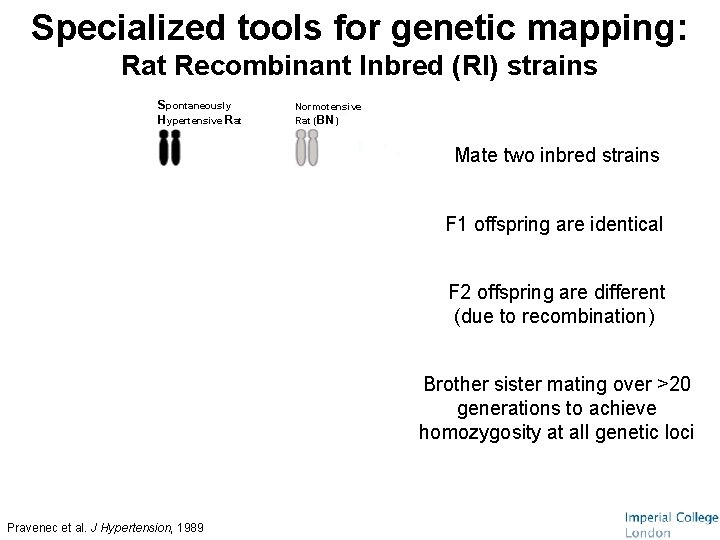
Specialized tools for genetic mapping: Rat Recombinant Inbred (RI) strains Spontaneously Hypertensive Rat Normotensive Rat (BN) Mate two inbred strains F 1 offspring are identical F 1 F 2 offspring are different (due to recombination) F 2 RI strains HXB 1 HXB 2 HXB 3 HXB 4 HXB 5 HXB 6 HXB 7 … Pravenec et al. J Hypertension, 1989 Brother sister mating over >20 generations to achieve homozygosity at all genetic loci

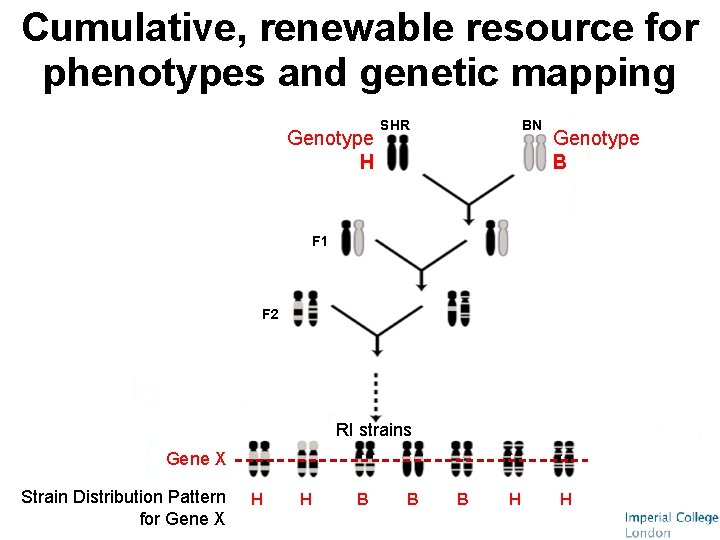
Cumulative, renewable resource for phenotypes and genetic mapping Genotype H SHR BN Genotype B F 1 F 2 RI strains Gene X Strain Distribution Pattern for Gene X H H B B B H H

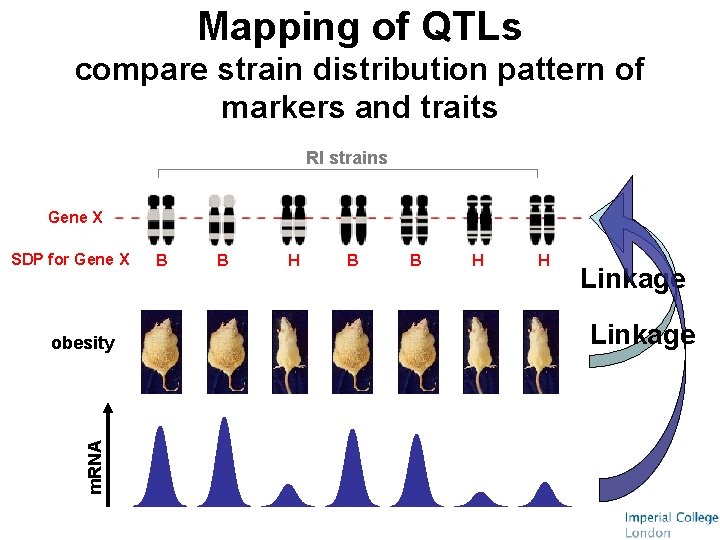
Mapping of QTLs compare strain distribution pattern of markers and traits RI strains Gene X SDP for Gene X m. RNA obesity B B H H Linkage

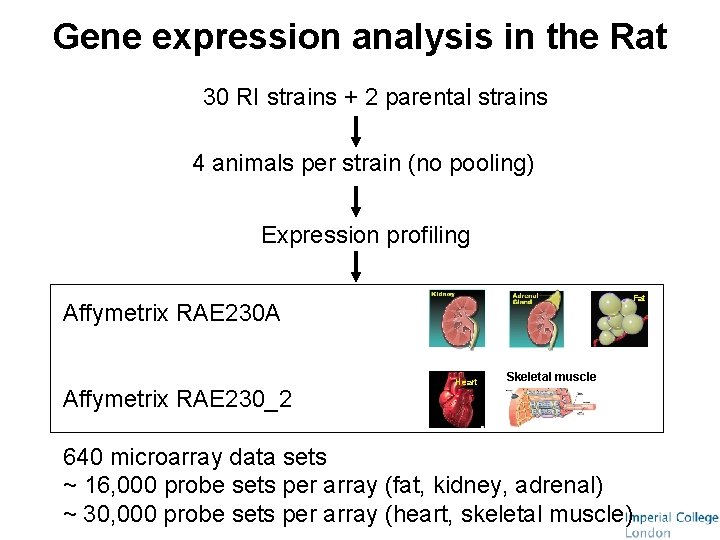
Gene expression analysis in the Rat 30 RI strains + 2 parental strains 4 animals per strain (no pooling) Expression profiling Fat Affymetrix RAE 230 A Affymetrix RAE 230_2 Heart Skeletal muscle 640 microarray data sets ~ 16, 000 probe sets per array (fat, kidney, adrenal) ~ 30, 000 probe sets per array (heart, skeletal muscle)

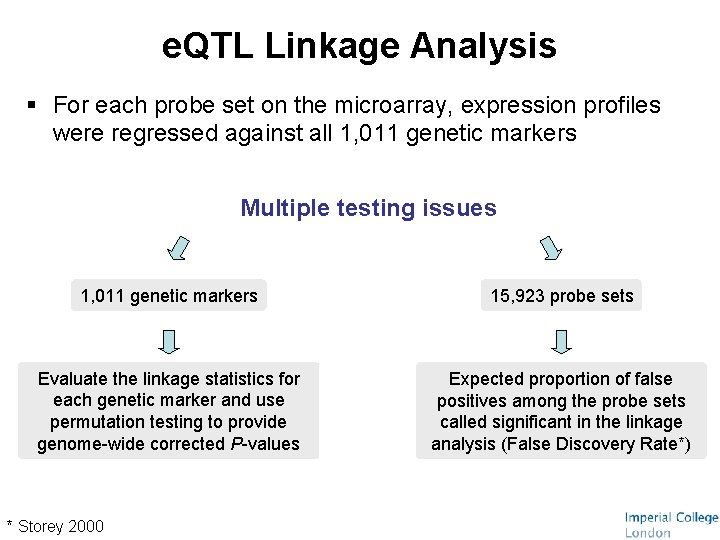
e. QTL Linkage Analysis § For each probe set on the microarray, expression profiles were regressed against all 1, 011 genetic markers Multiple testing issues 1, 011 genetic markers 15, 923 probe sets Evaluate the linkage statistics for each genetic marker and use permutation testing to provide genome-wide corrected P-values Expected proportion of false positives among the probe sets called significant in the linkage analysis (False Discovery Rate*) * Storey 2000

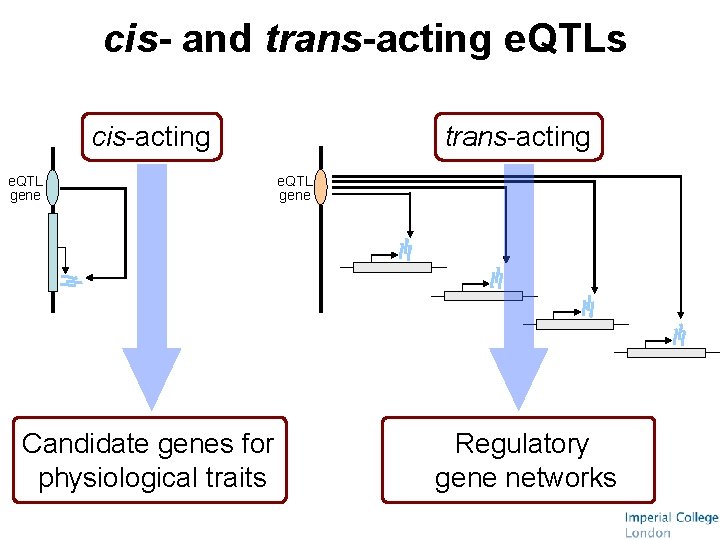
cis- and trans-acting e. QTLs cis-acting e. QTL gene Candidate genes for physiological traits trans-acting e. QTL gene Regulatory gene networks

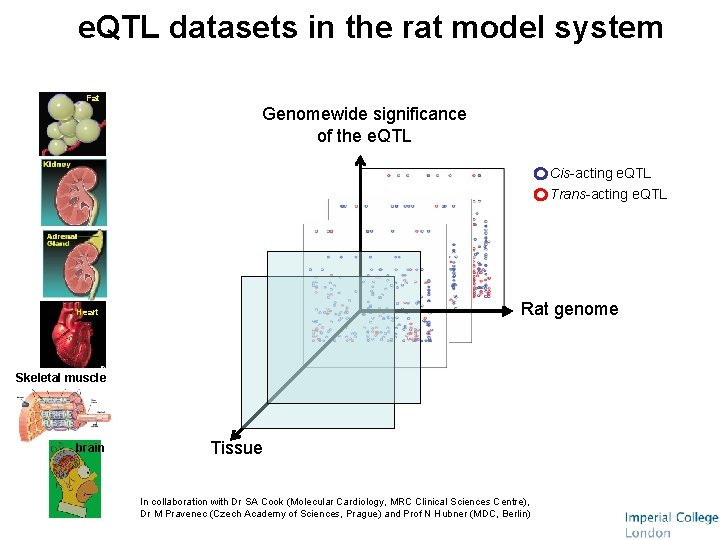
e. QTL datasets in the rat model system Fat Genomewide significance of the e. QTL Cis-acting e. QTL Trans-acting e. QTL Rat genome Heart Skeletal muscle brain Tissue In collaboration with Dr SA Cook (Molecular Cardiology, MRC Clinical Sciences Centre), Dr M Pravenec (Czech Academy of Sciences, Prague) and Prof N Hubner (MDC, Berlin)

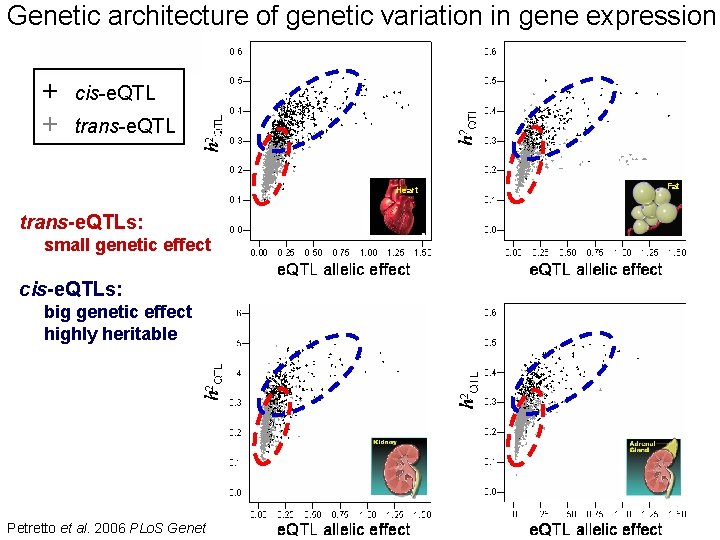
Genetic architecture of genetic variation in gene expression + + cis-e. QTL trans-e. QTL Heart trans-e. QTLs: small genetic effect cis-e. QTLs: big genetic effect highly heritable Petretto et al. 2006 PLo. S Genet Heart Fat

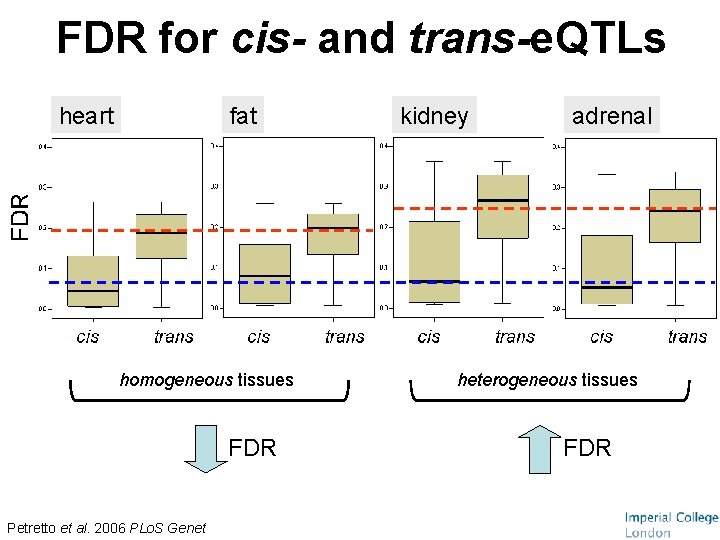
FDR for cis- and trans-e. QTLs heart fat homogeneous tissues FDR Petretto et al. 2006 PLo. S Genet kidney adrenal heterogeneous tissues FDR

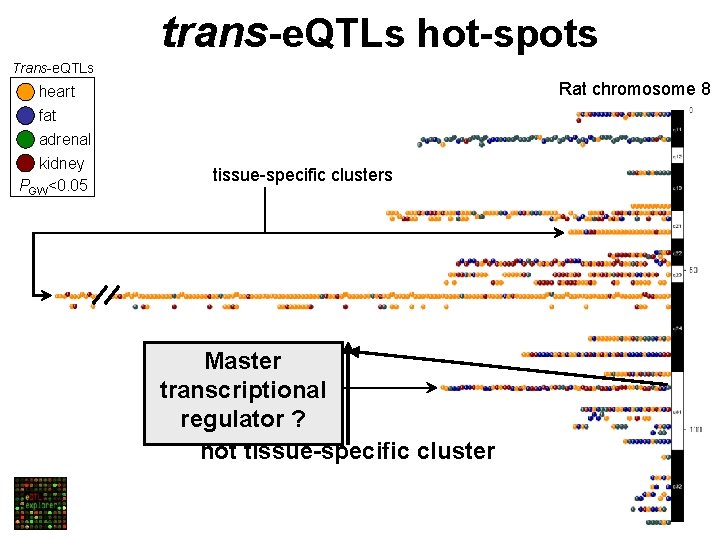
trans-e. QTLs hot-spots Trans-e. QTLs Rat chromosome 8 heart fat adrenal kidney PGW<0. 05 tissue-specific clusters Master transcriptional regulator ? not tissue-specific cluster

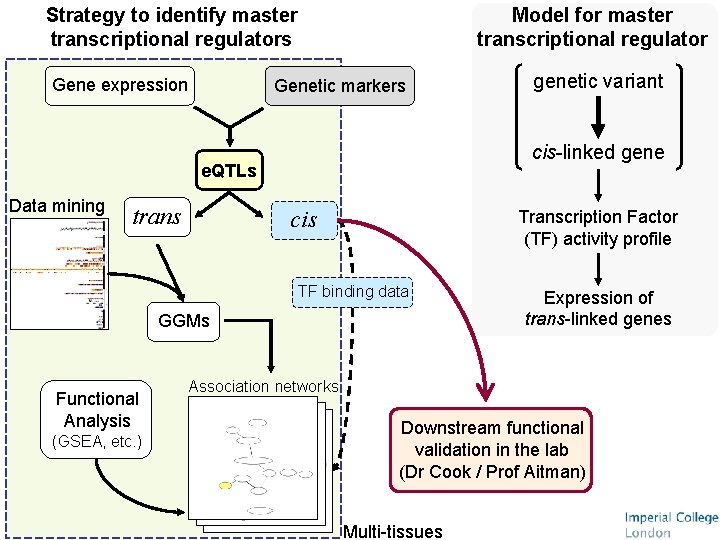
Strategy to identify master transcriptional regulators Gene expression Model for master transcriptional regulator Genetic markers cis-linked gene e. QTLs Data mining trans cis Transcription Factor (TF) activity profile TF binding data GGMs Functional Analysis (GSEA, etc. ) genetic variant Expression of trans-linked genes Association networks Downstream functional validation in the lab (Dr Cook / Prof Aitman) Multi-tissues

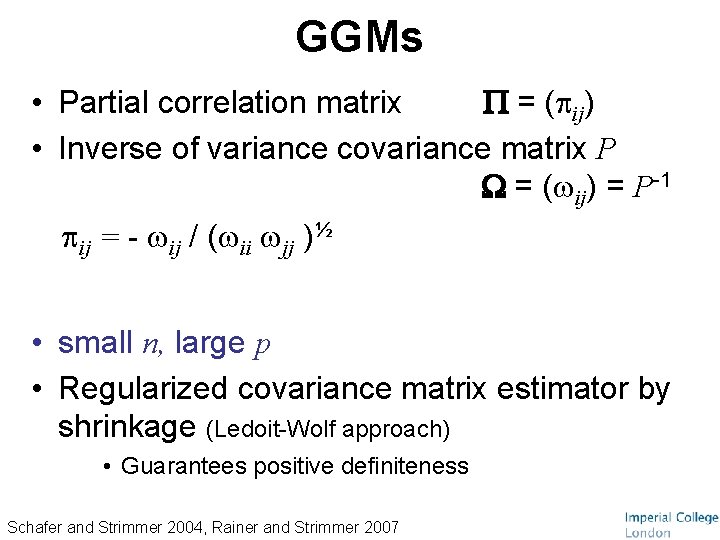
GGMs • Partial correlation matrix = ( ij) • Inverse of variance covariance matrix P = ( ij) = P-1 ij = - ij / ( ii jj )½ • small n, large p • Regularized covariance matrix estimator by shrinkage (Ledoit-Wolf approach) • Guarantees positive definiteness Schafer and Strimmer 2004, Rainer and Strimmer 2007

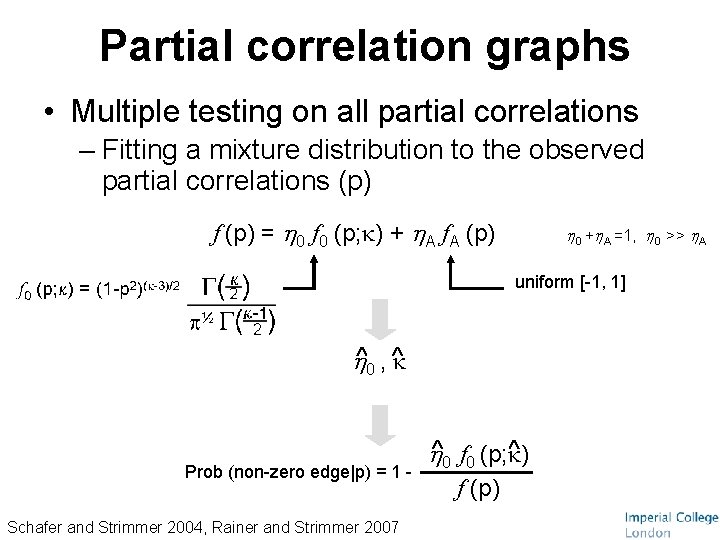
Partial correlation graphs • Multiple testing on all partial correlations – Fitting a mixture distribution to the observed partial correlations (p) f (p) = 0 f 0 (p; ) + A f. A (p) 0 + A =1, 0 >> A uniform [-1, 1] 0 , Prob (non-zero edge|p) = 1 Schafer and Strimmer 2004, Rainer and Strimmer 2007 0 f 0 (p; ) f (p)

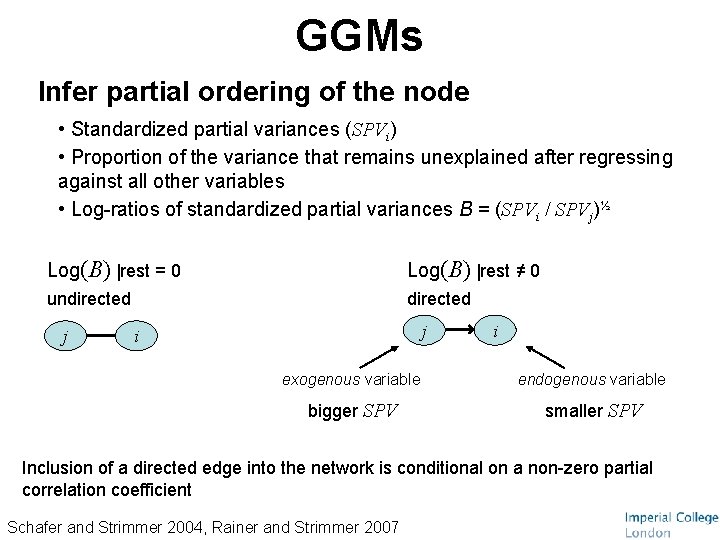
GGMs Infer partial ordering of the node • Standardized partial variances (SPVi) • Proportion of the variance that remains unexplained after regressing against all other variables • Log-ratios of standardized partial variances B = (SPVi / SPVj)½ Log(B) |rest = 0 Log(B) |rest ≠ 0 undirected j j i i exogenous variable endogenous variable bigger SPV smaller SPV Inclusion of a directed edge into the network is conditional on a non-zero partial correlation coefficient Schafer and Strimmer 2004, Rainer and Strimmer 2007

Hypothesis driven analysis 1. Gene expression levels under genetic control (i. e. , ‘structural’ genetic perturbation) 2. Co-expression of trans-e. QTLs point to common regulation by a single gene Graphical Gaussian models • Detect conditionally dependent trans-e. QTL genes • Infer partial ordering of the nodes (directed edges)

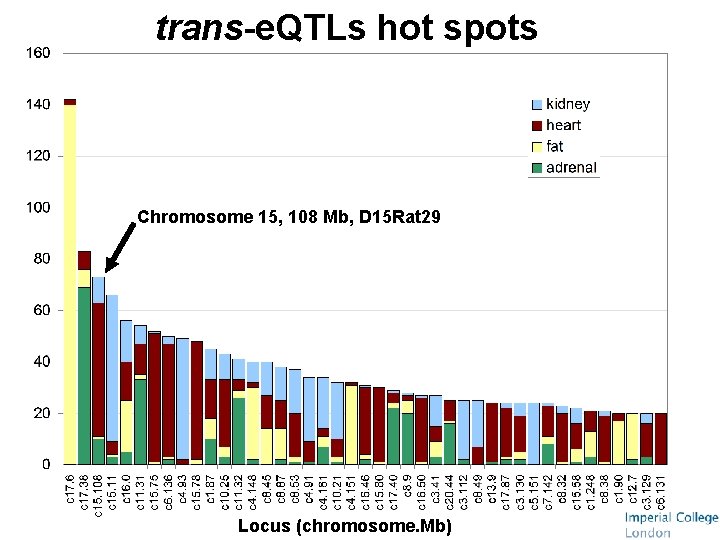
trans-e. QTLs hot spots Chromosome 15, 108 Mb, D 15 Rat 29 Locus (chromosome. Mb)

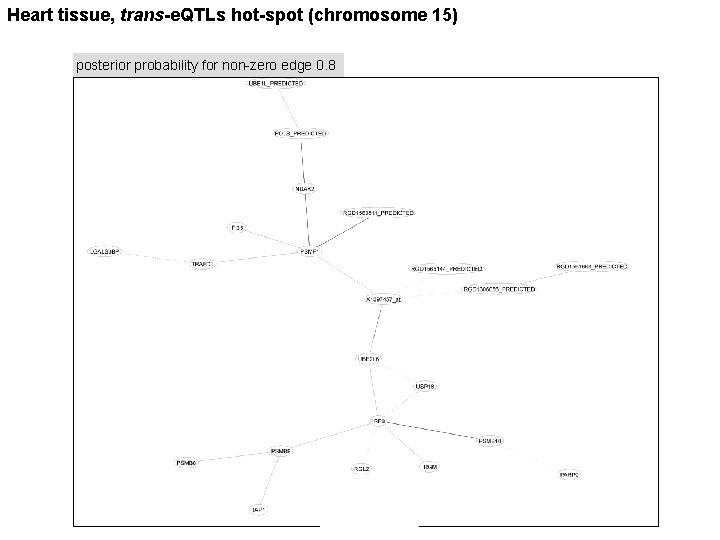
Heart tissue, trans-e. QTLs hot-spot (chromosome 15) posterior probability for non-zero edge 0. 8

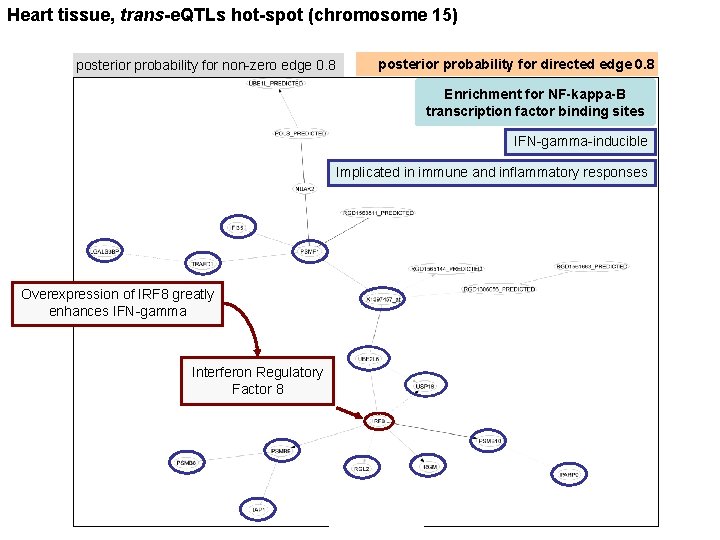
Heart tissue, trans-e. QTLs hot-spot (chromosome 15) posterior probability for non-zero edge 0. 8 posterior probability for directed edge 0. 8 Enrichment for NF-kappa-B transcription factor binding sites IFN-gamma-inducible Implicated in immune and inflammatory responses Overexpression of IRF 8 greatly enhances IFN-gamma Interferon Regulatory Factor 8

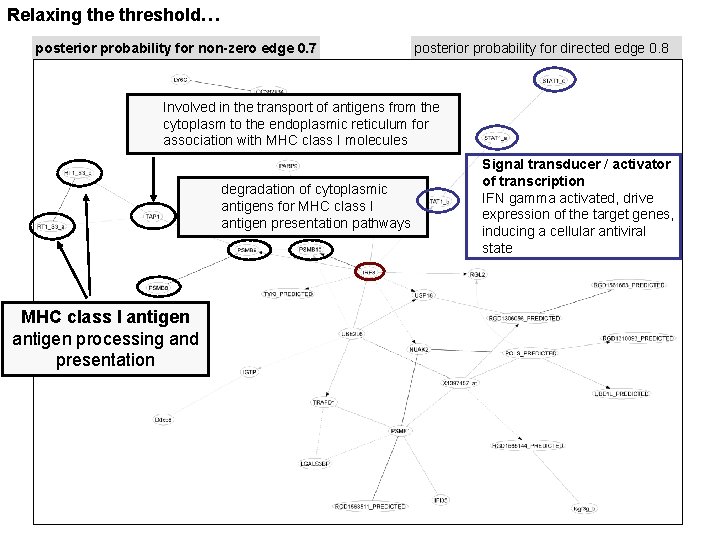
Relaxing the threshold… posterior probability for non-zero edge 0. 7 posterior probability for directed edge 0. 8 Involved in the transport of antigens from the cytoplasm to the endoplasmic reticulum for association with MHC class I molecules degradation of cytoplasmic antigens for MHC class I antigen presentation pathways MHC class I antigen processing and presentation Signal transducer / activator of transcription IFN gamma activated, drive expression of the target genes, inducing a cellular antiviral state

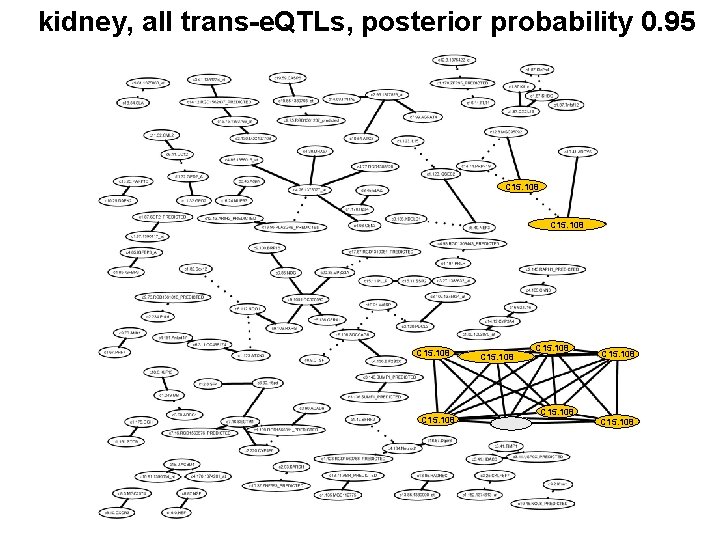
Is this association graph tissue specific?

kidney, all trans-e. QTLs, posterior probability 0. 95 C 15. 108 C 15. 108

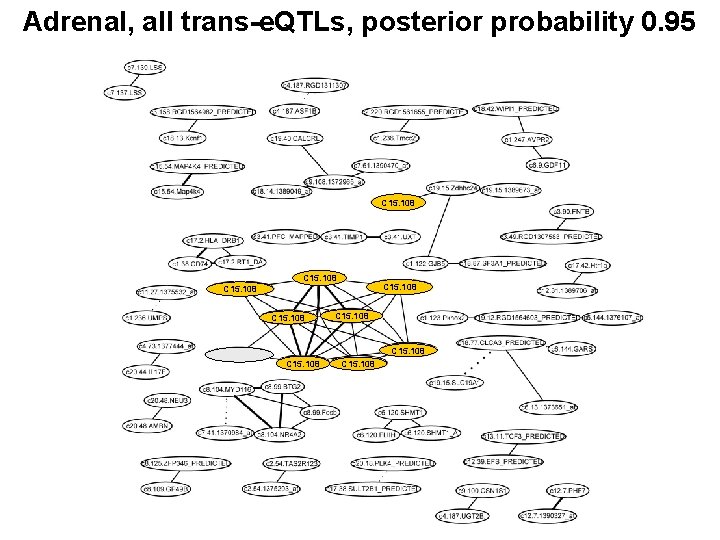
Adrenal, all trans-e. QTLs, posterior probability 0. 95 C 15. 108 C 15. 108

FC in the parental strains adrenal heart kidney Microarray data: dysregulated genes IRF - transcription factor inflammatory response FC in RI strains interferon-stimulated transcription factor type I interferon (IFN) inducible gene Trans-e. QTL genes detected in multiple tissues

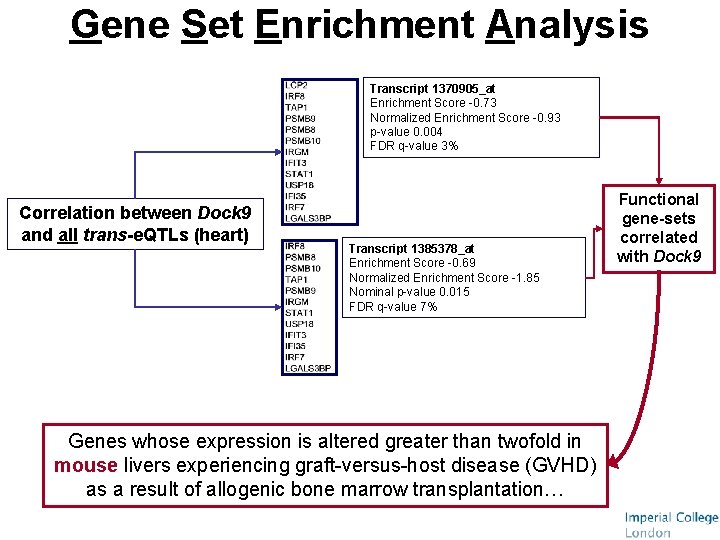
Model for master transcriptional regulator cis-acting e. QTLs within the cluster region Transcripts representing Dock 9 genetic variant cis-linked gene Transcription Factor (TF) activity profile Expression of trans-linked genes Trans cluster Cis e. QTLs Pearson Correlation 100, 000 permutations Bonferroni corrected

Gene Set Enrichment Analysis Transcript 1370905_at Enrichment Score -0. 73 Normalized Enrichment Score -0. 93 p-value 0. 004 FDR q-value 3% Correlation between Dock 9 and all trans-e. QTLs (heart) Transcript 1385378_at Enrichment Score -0. 69 Normalized Enrichment Score -1. 85 Nominal p-value 0. 015 FDR q-value 7% Genes whose expression is altered greater than twofold in mouse livers experiencing graft-versus-host disease (GVHD) as a result of allogenic bone marrow transplantation… Functional gene-sets correlated with Dock 9

Other examples

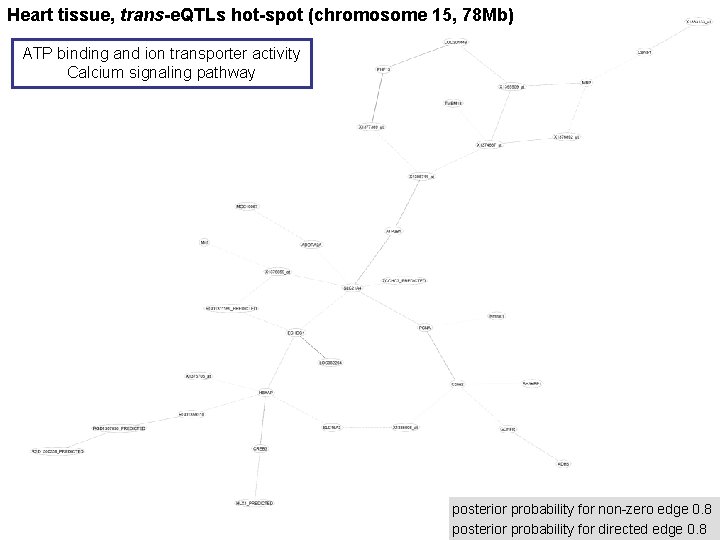
Heart tissue, trans-e. QTLs hot-spot (chromosome 15, 78 Mb) ATP binding and ion transporter activity Calcium signaling pathway posterior probability for non-zero edge 0. 8 posterior probability for directed edge 0. 8

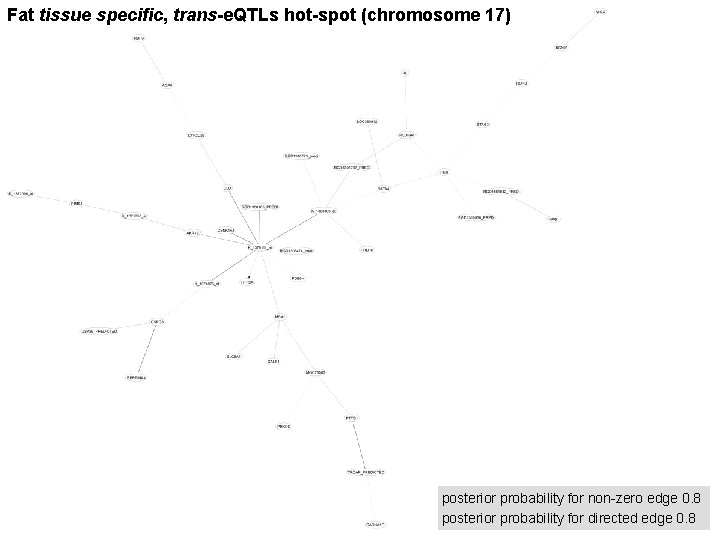
Fat tissue specific, trans-e. QTLs hot-spot (chromosome 17) posterior probability for non-zero edge 0. 8 posterior probability for directed edge 0. 8

Summary • Genome-wide e. QTL data provide new insights into gene regulatory networks • GGMs applied to trans-e. QTL hotspots identified dysregulated pathway related to inflammation • Hypothesis-driven inference can be a powerful approach to dissect regulatory networks

Acknowledgments Sylvia Richardson Tim Aitman Stuart Cook Jonathan Mangion Rizwan Sarwar collaborators: Norbert Hubner (MDC, Berlin) Michael Pravenec (Institute of Physiology, Prague)

Extra slides

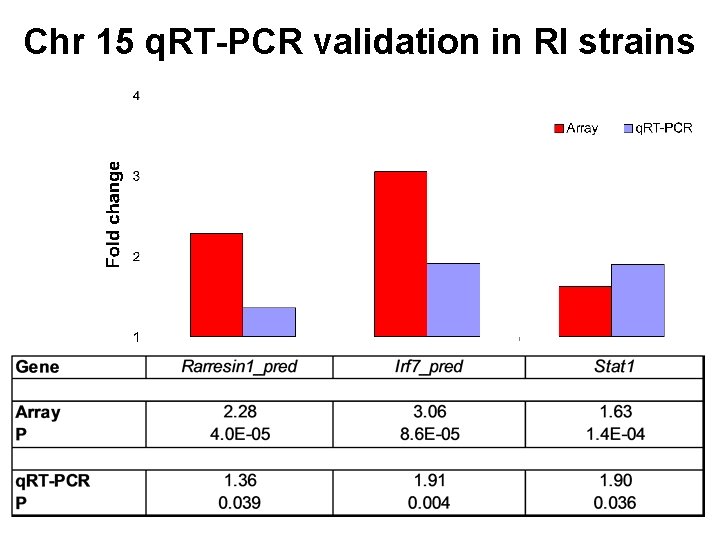
Chr 15 q. RT-PCR validation in RI strains

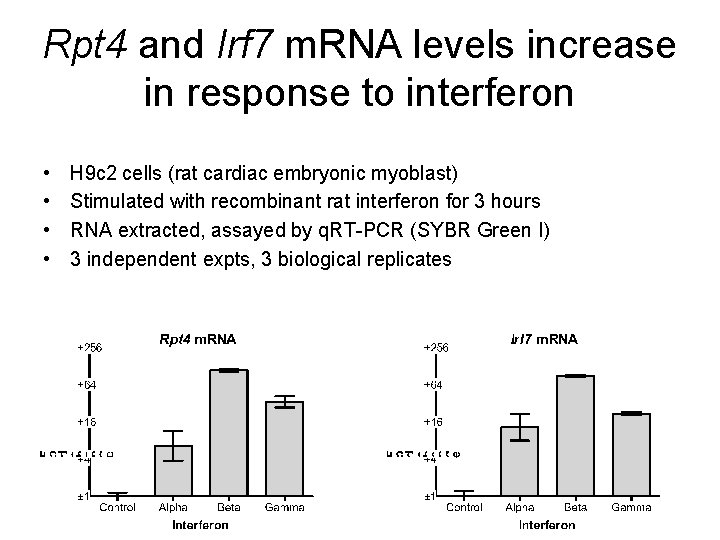
Rpt 4 and Irf 7 m. RNA levels increase in response to interferon • • H 9 c 2 cells (rat cardiac embryonic myoblast) Stimulated with recombinant rat interferon for 3 hours RNA extracted, assayed by q. RT-PCR (SYBR Green I) 3 independent expts, 3 biological replicates