Insights into lipid structure and dynamics some neutron




- Slides: 46

Insights into lipid structure and dynamics: some neutron scattering case studies Paul Butler CHEG 867 SCATTERING METHODS

What can you study with neutrons? Shear thickening fluids Superconductivity Marbles Geological formations Polymer composites Self assembled systems Explosives Protein complexes Magnetic nanoparticles Hydrogen storage materials Pressure vessel steels Monoclonal Antibodies Colloid Physics Etc. Center for Neutron Sciences

When is it useful? Not the best Survey instrument – not very good for unknowns. But … can be very powerful tools, particularly when using deuteration, for quantitatively answering questions about structure (size, shape, orientation) or dynamics: • Pose the question clearly (propose hypothesis) • Carefully design experiment • Careful analysis Center for Neutron Sciences

Neutrons don’t lie (tales from the crypt) Wrong solvent Wrong cell materials Unstable sample Impurities Quality of sample (monodisperse, spheres, etc. ) Center for Neutron Sciences

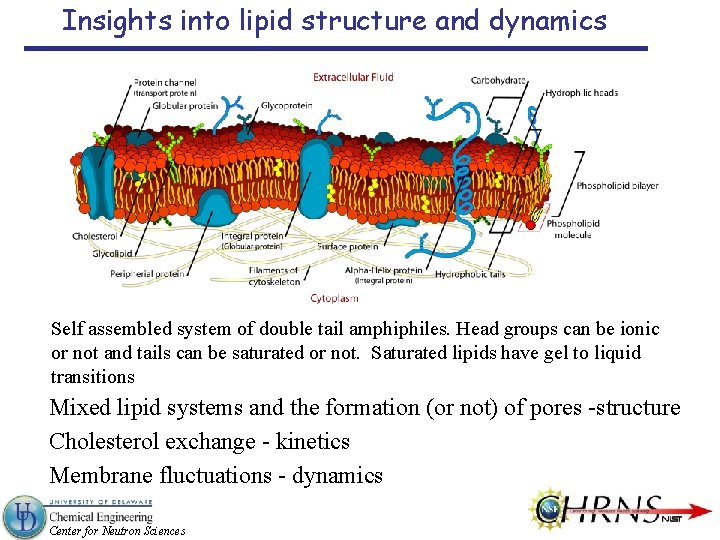
Insights into lipid structure and dynamics Self assembled system of double tail amphiphiles. Head groups can be ionic or not and tails can be saturated or not. Saturated lipids have gel to liquid transitions Mixed lipid systems and the formation (or not) of pores -structure Cholesterol exchange - kinetics Membrane fluctuations - dynamics Center for Neutron Sciences

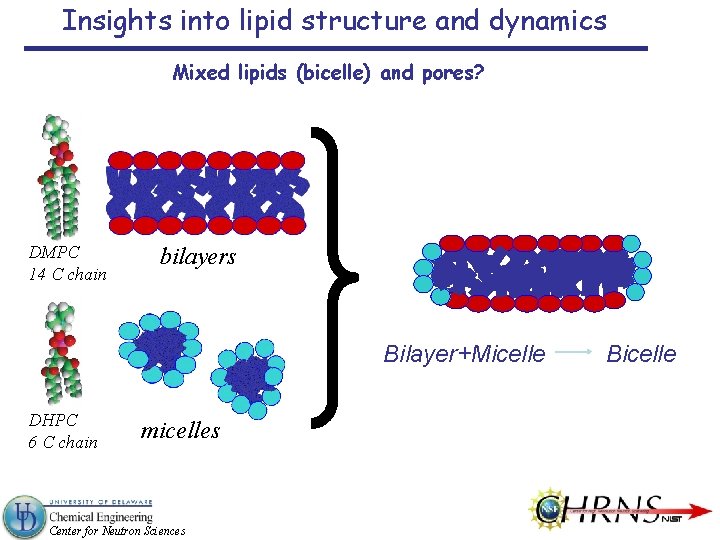
Insights into lipid structure and dynamics Mixed lipids (bicelle) and pores? DMPC 14 C chain bilayers Bilayer+Micelle DHPC 6 C chain micelles Center for Neutron Sciences Bicelle

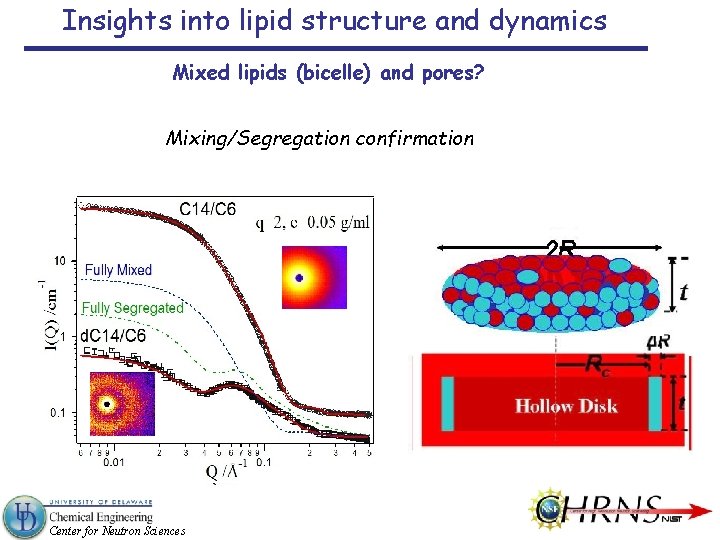
Insights into lipid structure and dynamics Mixed lipids (bicelle) and pores? Mixing/Segregation confirmation Center for Neutron Sciences

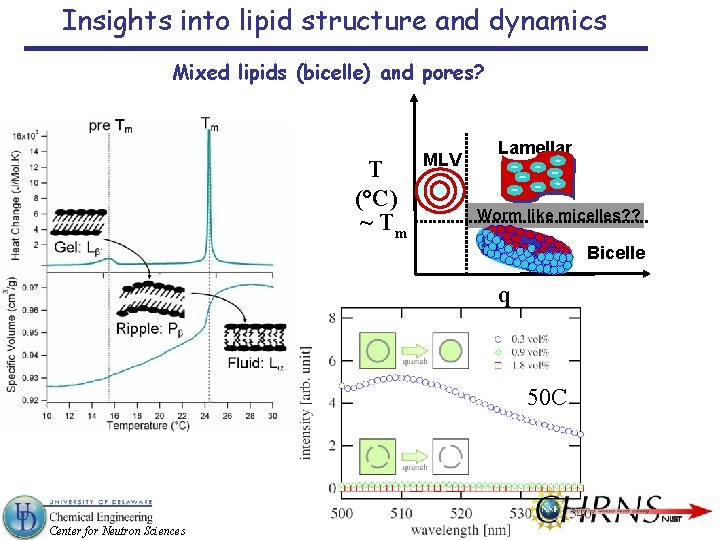
Insights into lipid structure and dynamics Mixed lipids (bicelle) and pores? T (°C) ~ Tm MLV Lamellar Worm like micelles? ? Bicelle q 50 C Center for Neutron Sciences

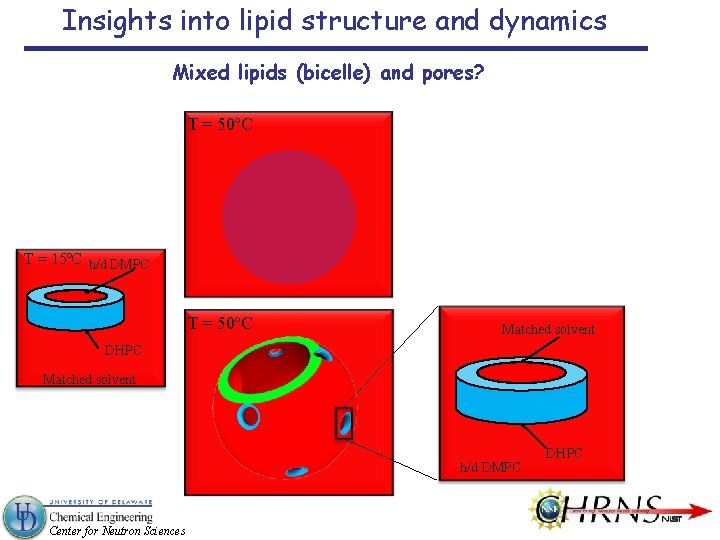
Insights into lipid structure and dynamics Mixed lipids (bicelle) and pores? T = 50ºC T = 15ºC h/d DMPC T = 50ºC Matched solvent DHPC Matched solvent h/d DMPC Center for Neutron Sciences DHPC

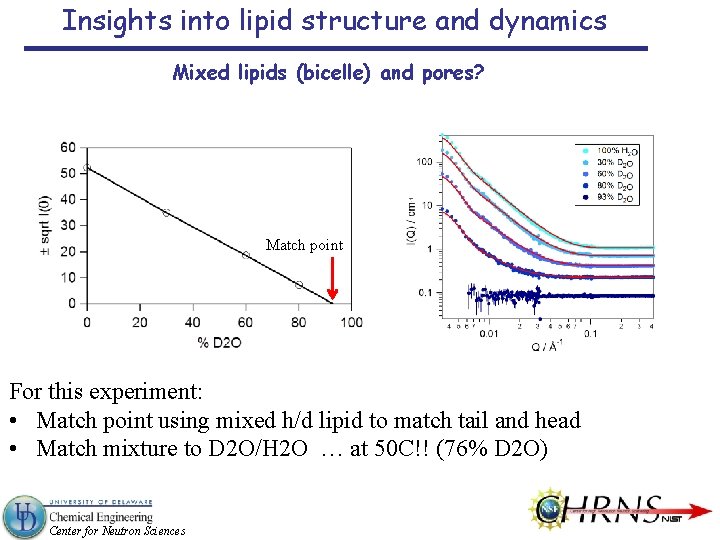
Insights into lipid structure and dynamics Mixed lipids (bicelle) and pores? Match point For this experiment: • Match point using mixed h/d lipid to match tail and head • Match mixture to D 2 O/H 2 O … at 50 C!! (76% D 2 O) Center for Neutron Sciences

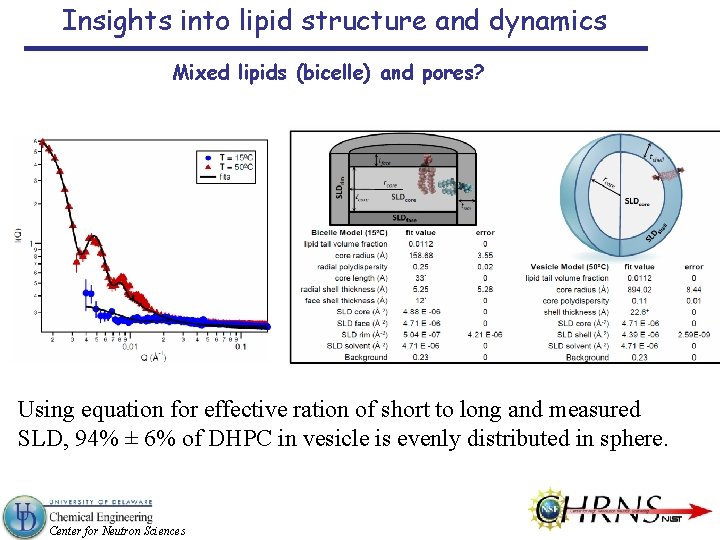
Insights into lipid structure and dynamics Mixed lipids (bicelle) and pores? Using equation for effective ration of short to long and measured SLD, 94% ± 6% of DHPC in vesicle is evenly distributed in sphere. Center for Neutron Sciences

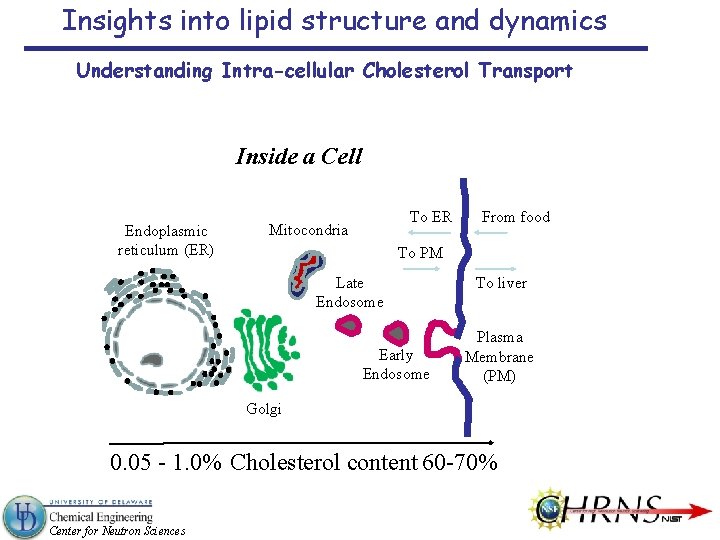
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Inside a Cell Endoplasmic reticulum (ER) To ER Mitocondria From food To PM Late Endosome Early Endosome To liver Plasma Membrane (PM) Golgi 0. 05 - 1. 0% Cholesterol content 60 -70% Center for Neutron Sciences

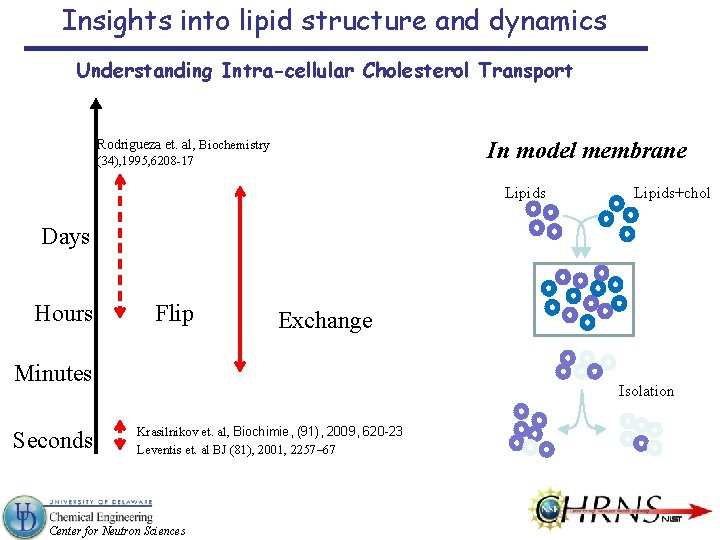
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport In model membrane Rodrigueza et. al, Biochemistry (34), 1995, 6208 -17 Lipids+chol Days Hours Flip Exchange Minutes Seconds Isolation Krasilnikov et. al, Biochimie, (91), 2009, 620 -23 Leventis et. al BJ (81), 2001, 2257– 67 Center for Neutron Sciences

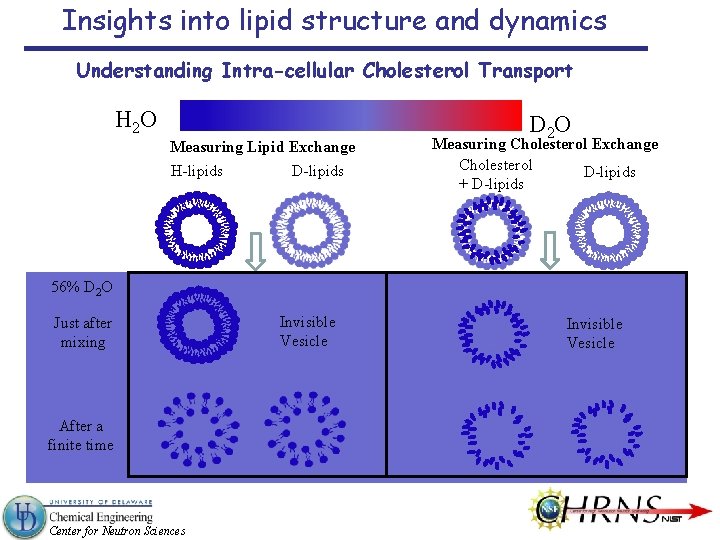
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport H 2 O Measuring Lipid Exchange H-lipids D 2 O Measuring Cholesterol Exchange Cholesterol D-lipids + D-lipids 56% D 2 O Just after mixing After a finite time Center for Neutron Sciences Invisible Vesicle

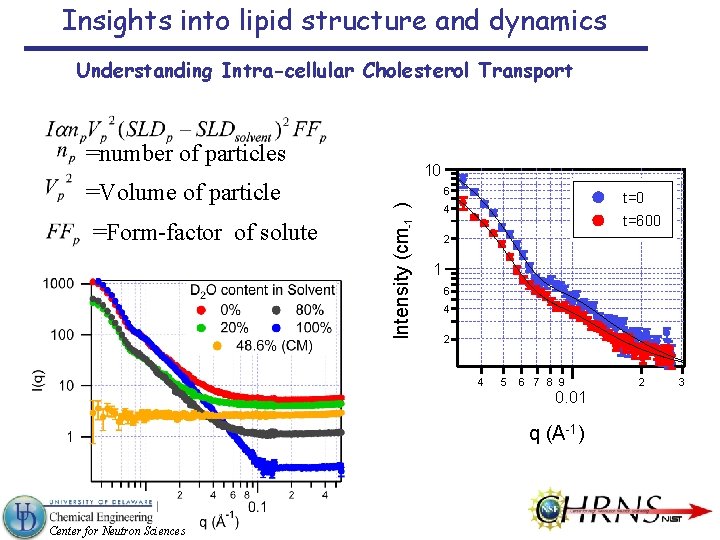
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport =number of particles =Form-factor of solute 6 Intensity (cm-1 ) =Volume of particle 10 t=0 4 t=600 2 1 6 4 2 4 5 6 7 8 9 0. 01 q (A-1) Center for Neutron Sciences 2 3

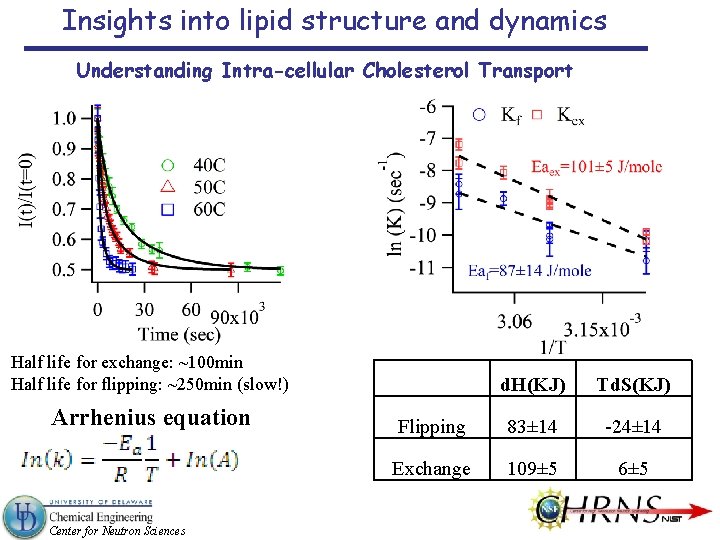
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Half life for exchange: ~100 min Half life for flipping: ~250 min (slow!) Arrhenius equation Center for Neutron Sciences d. H(KJ) Td. S(KJ) Flipping 83± 14 -24± 14 Exchange 109± 5 6± 5

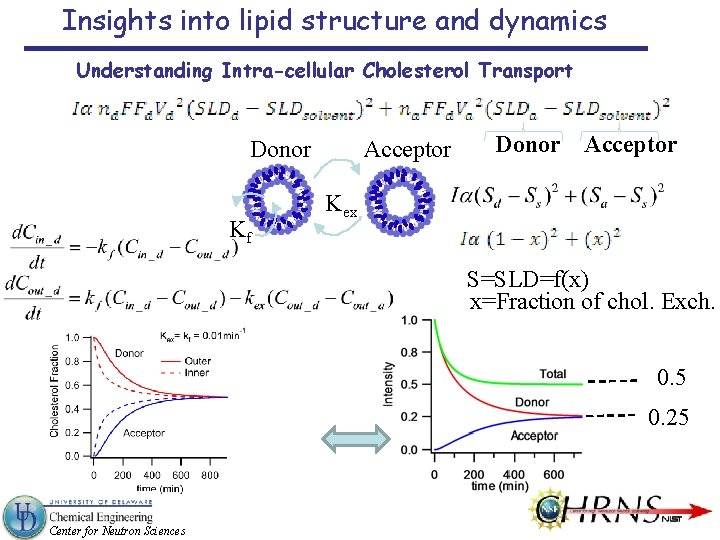
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Donor Kf Acceptor Donor Acceptor Kex S=SLD=f(x) x=Fraction of chol. Exch. 0. 5 0. 25 Center for Neutron Sciences

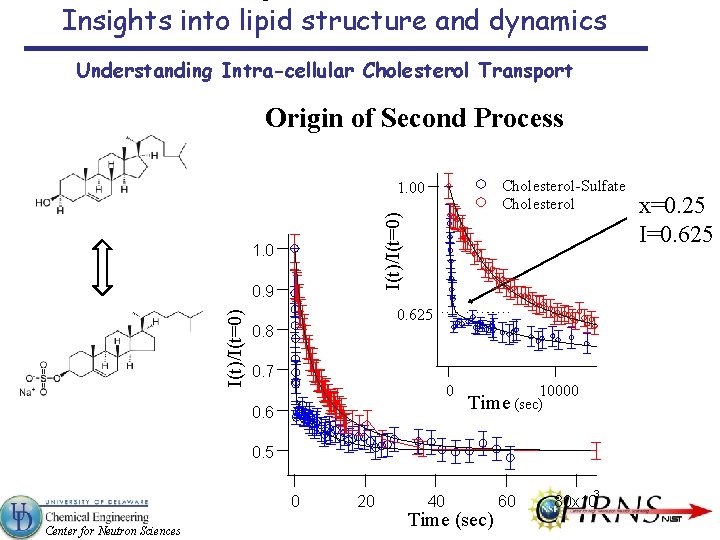
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Origin of Second Process Cholesterol-Sulfate Cholesterol I(t)/I(t=0) 1. 00 1. 0 I(t)/I(t=0) 0. 9 0. 625 0. 8 0. 7 0 0. 6 10000 Time (sec) 0. 5 0 Center for Neutron Sciences 20 40 Time (sec) 60 3 80 x 10 x=0. 25 I=0. 625

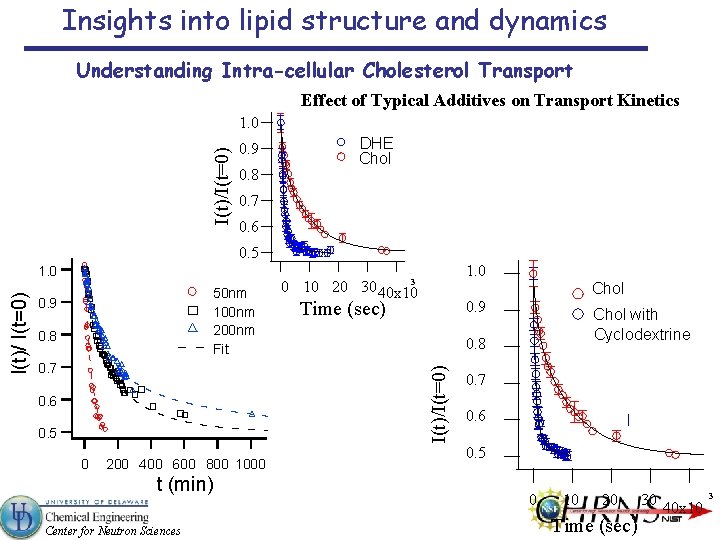
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Effect of Typical Additives on Transport Kinetics I(t)/I(t=0) 1. 0 0. 9 0. 8 DHE Chol 0. 7 0. 6 0. 5 50 nm 100 nm 200 nm Fit 0. 9 0. 8 0. 7 1. 0 3 0 10 20 30 40 x 10 Chol Time (sec) 0. 9 0. 6 0. 5 0 200 400 600 800 1000 t (min) Center for Neutron Sciences Chol with Cyclodextrine 0. 8 I(t)/I(t=0) I(t)/ I(t=0) 1. 0 0. 7 0. 6 0. 5 0 10 20 Time (sec) 30 40 x 10 3

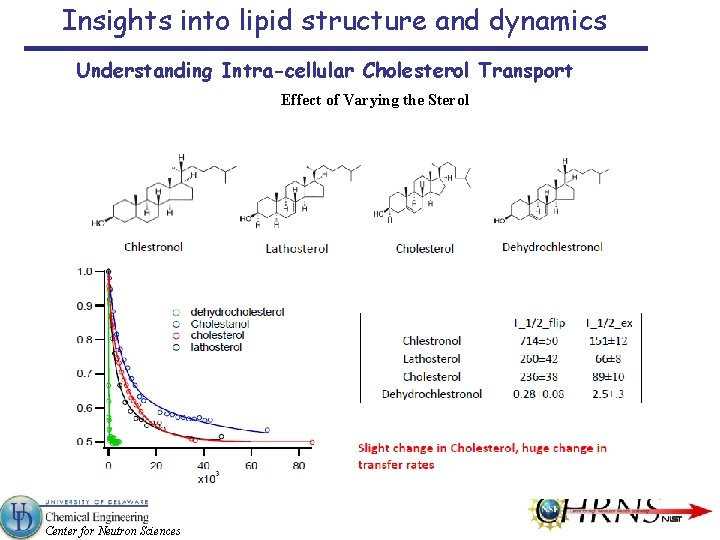
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Effect of Varying the Sterol Center for Neutron Sciences

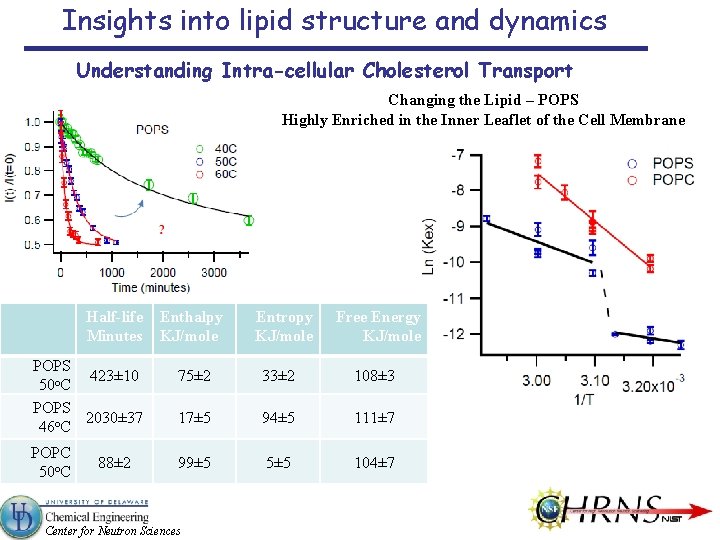
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Changing the Lipid – POPS Highly Enriched in the Inner Leaflet of the Cell Membrane Half-life Minutes POPS 423± 10 50 o. C POPS 2030± 37 46 o. C POPC 50 o. C 88± 2 Enthalpy KJ/mole Entropy KJ/mole Free Energy KJ/mole 75± 2 33± 2 108± 3 17± 5 94± 5 111± 7 99± 5 5± 5 104± 7 Center for Neutron Sciences

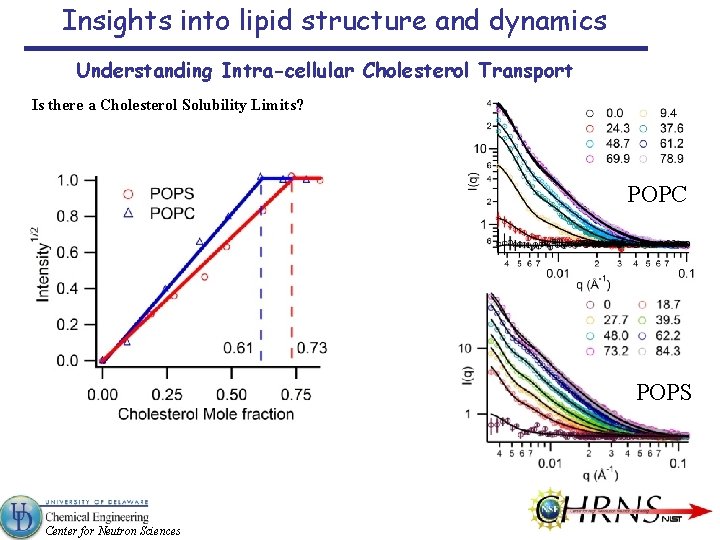
Insights into lipid structure and dynamics Understanding Intra-cellular Cholesterol Transport Is there a Cholesterol Solubility Limits? POPC POPS Center for Neutron Sciences

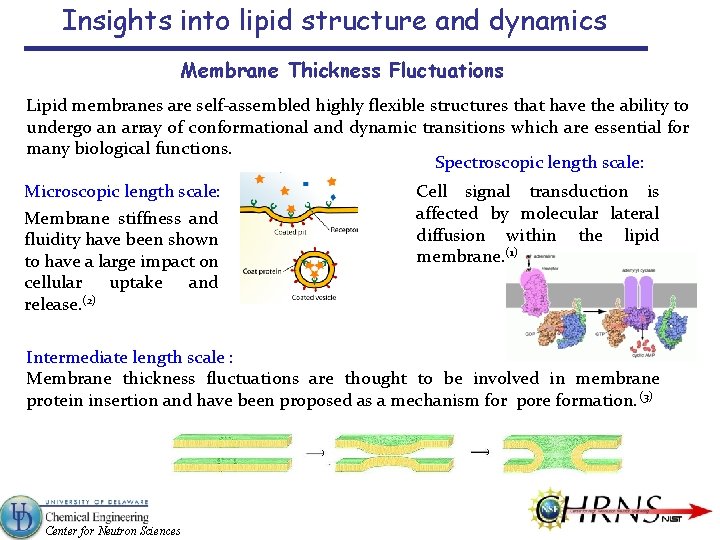
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Lipid membranes are self-assembled highly flexible structures that have the ability to undergo an array of conformational and dynamic transitions which are essential for many biological functions. Spectroscopic length scale: Microscopic length scale: Membrane stiffness and fluidity have been shown to have a large impact on cellular uptake and (2) release. Cell signal transduction is affected by molecular lateral diffusion within the lipid membrane. (1) Intermediate length scale : Membrane thickness fluctuations are thought to be involved in membrane protein insertion and have been proposed as a mechanism for pore formation. (3) Center for Neutron Sciences

Insights into lipid structure and dynamics Membrane Thickness Fluctuations Breathing model of a lipid bilayer by Miller THEORY Miller, Top. Bioelectrochem. Bioenerg. 4, 161 (1981). ; Bach and Miller, Biophys. J. 29, 183 (1980). ; Miller, Biophys. J. 45, 643 (1984). Amplitude of the fluctuations reaches ≈ 15 Å or more from the geometrical constraints (volume conservation) Hladky and Gruen, Biophys. J. 38, 251 Thickness fluctuations by Hladky and Gruen (1982). Thickness fluctuations occur, but the amplitude is small. Long wavelength fluctuation amplitude is negligible Short wavelength fluctuations (< 30 Å) are severely limited Intermediate wavelength fluctuation amplitude < 10 Å Deformation free energy of bilayer membranes by Huang Center for Neutron Sciences Huang, Biophys. J. 50, 1061 (1986).

Insights into lipid structure and dynamics Membrane Thickness Fluctuations SIMULATION Lindahl and Edholm, Biophys. J. 79, 426 (2000). Simulation time scale: 10 or 60 ns Monolayer RMS amplitude for peristaltic (thickness fluctuation): 2. 5 Å Suppressed thickness fluctuations in the gel phase West and Schmid, Soft Matter 6, 1275 (2010). Thickness fluctuation amplitude is much smaller than that in the fluid phase Center for Neutron Sciences

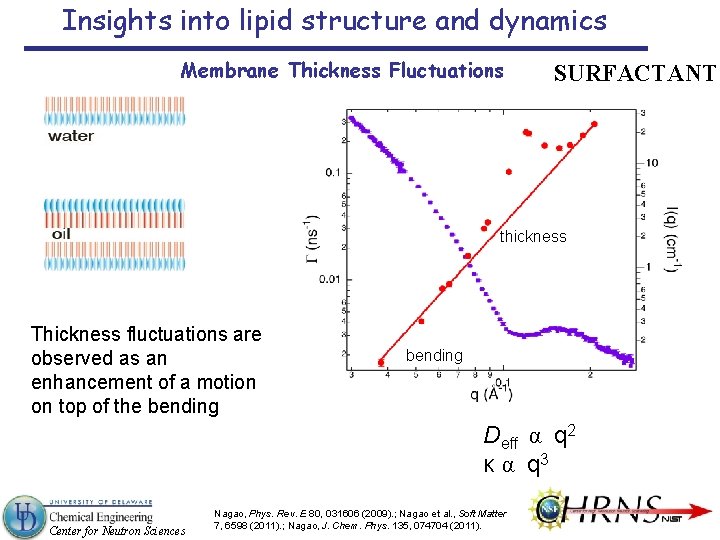
Insights into lipid structure and dynamics Membrane Thickness Fluctuations SURFACTANT thickness Thickness fluctuations are observed as an enhancement of a motion on top of the bending Deff α q 2 κ α q 3 Center for Neutron Sciences Nagao, Phys. Rev. E 80, 031606 (2009). ; Nagao et al. , Soft Matter 7, 6598 (2011). ; Nagao, J. Chem. Phys. 135, 074704 (2011).

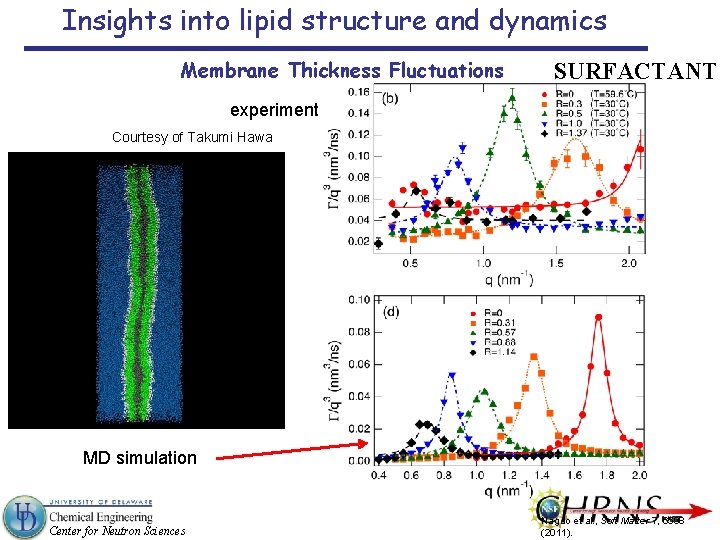
Insights into lipid structure and dynamics Membrane Thickness Fluctuations SURFACTANT experiment Courtesy of Takumi Hawa MD simulation Center for Neutron Sciences Nagao et al. , Soft Matter 7, 6598 (2011).

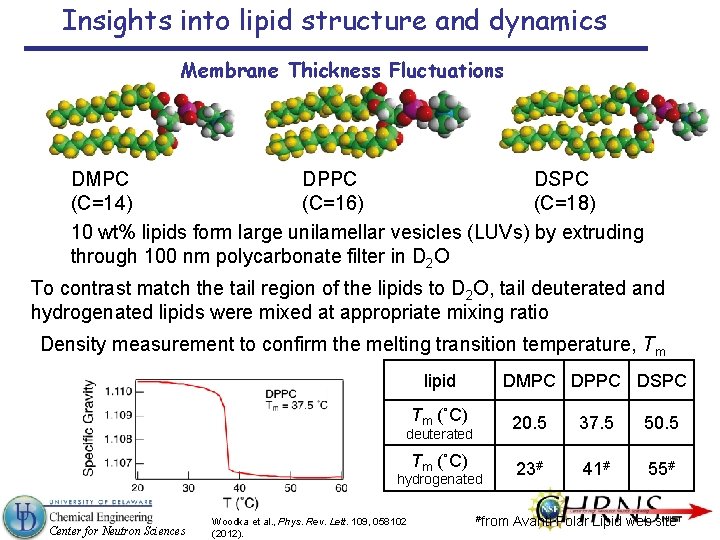
Insights into lipid structure and dynamics Membrane Thickness Fluctuations DMPC DPPC DSPC (C=14) (C=16) (C=18) 10 wt% lipids form large unilamellar vesicles (LUVs) by extruding through 100 nm polycarbonate filter in D 2 O To contrast match the tail region of the lipids to D 2 O, tail deuterated and hydrogenated lipids were mixed at appropriate mixing ratio Density measurement to confirm the melting transition temperature, Tm lipid DMPC DPPC DSPC Tm (˚C) 20. 5 37. 5 50. 5 Tm (˚C) 23# 41# 55# deuterated hydrogenated Center for Neutron Sciences Woodka et al. , Phys. Rev. Lett. 109, 058102 (2012). #from Avanti Polar Lipid web site

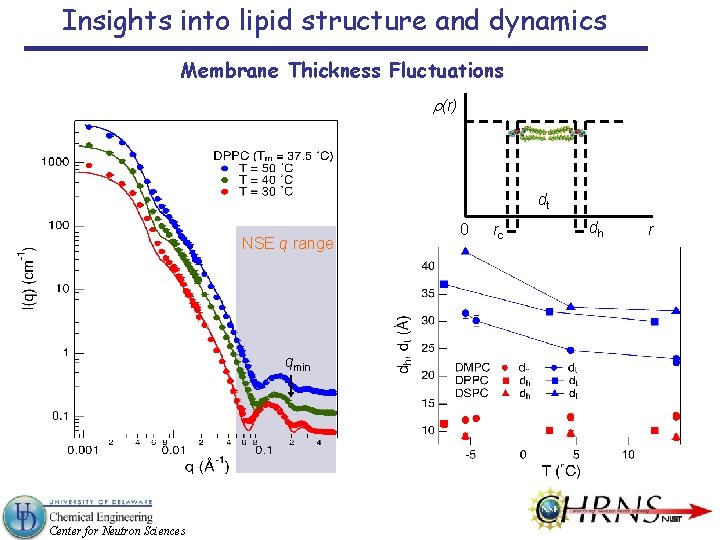
Insights into lipid structure and dynamics Membrane Thickness Fluctuations r(r) dt NSE q range qmin Center for Neutron Sciences 0 rc dh r

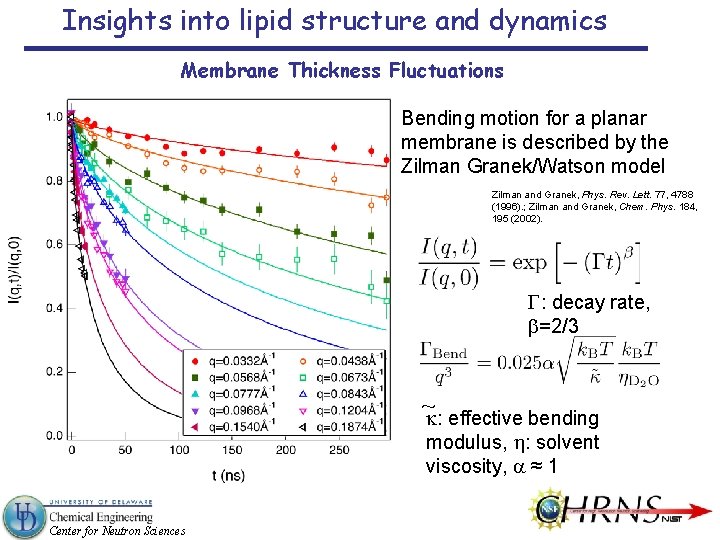
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Bending motion for a planar membrane is described by the Zilman Granek/Watson model Zilman and Granek, Phys. Rev. Lett. 77, 4788 (1996). ; Zilman and Granek, Chem. Phys. 184, 195 (2002). G: decay rate, b=2/3 ~k: effective bending modulus, h: solvent viscosity, a ≈ 1 Center for Neutron Sciences

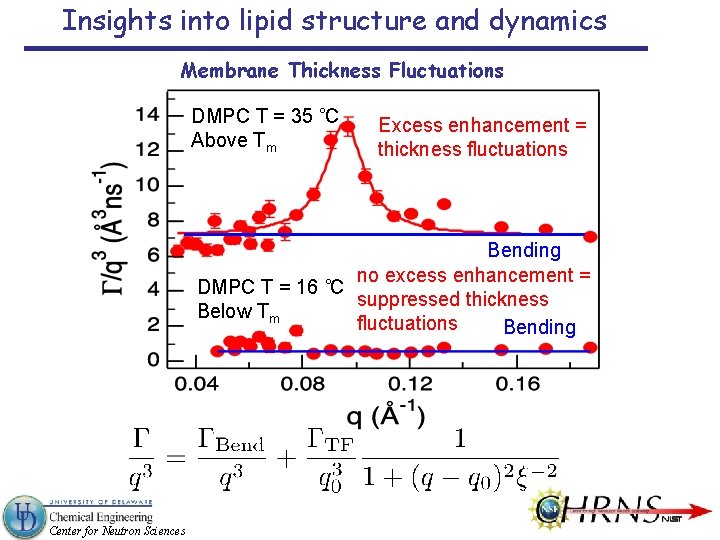
Insights into lipid structure and dynamics Membrane Thickness Fluctuations DMPC T = 35 ˚C Above Tm Excess enhancement = thickness fluctuations Bending no excess enhancement = DMPC T = 16 ˚C suppressed thickness Below Tm fluctuations Bending Center for Neutron Sciences

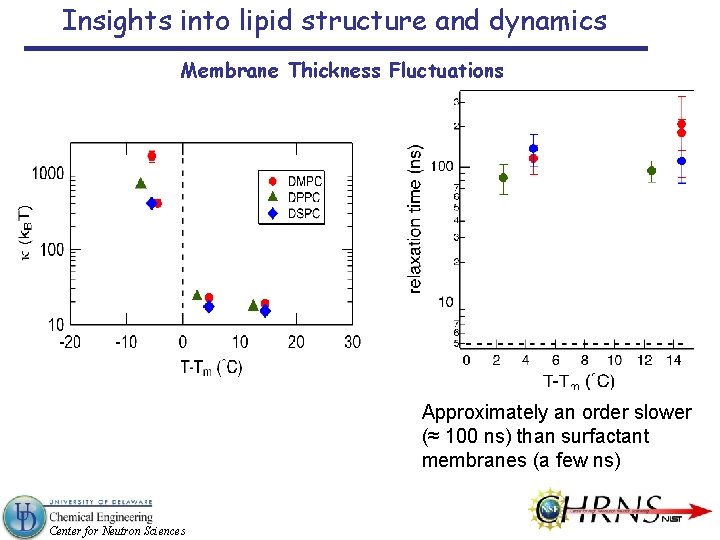
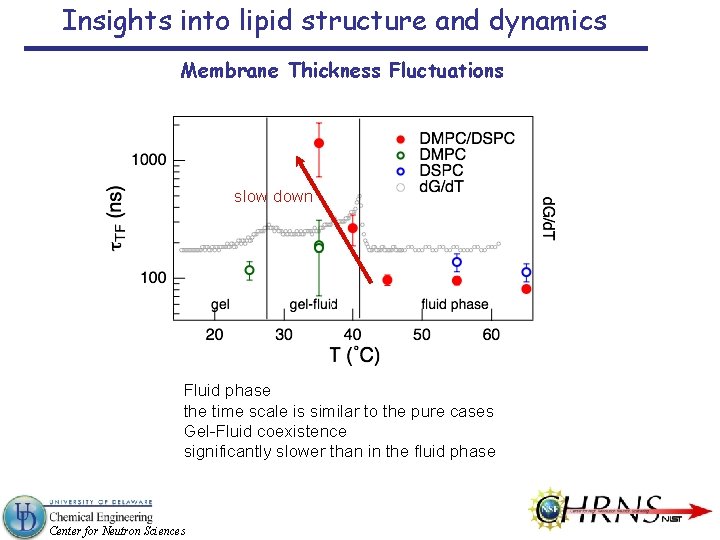
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Approximately an order slower (≈ 100 ns) than surfactant membranes (a few ns) Center for Neutron Sciences

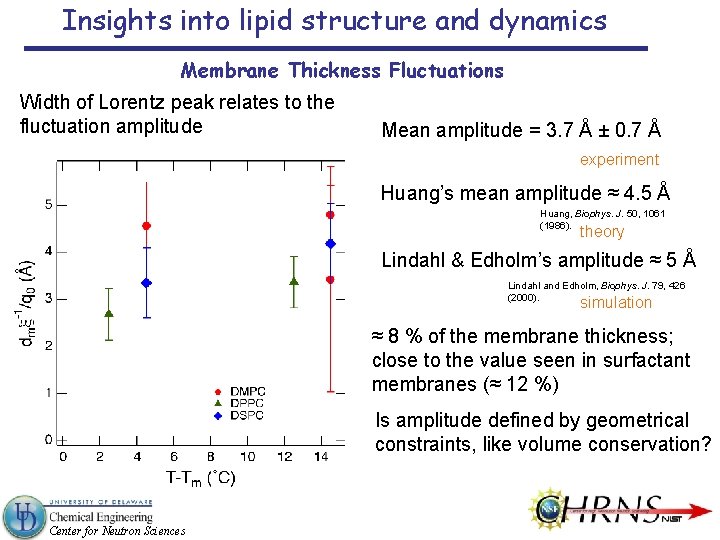
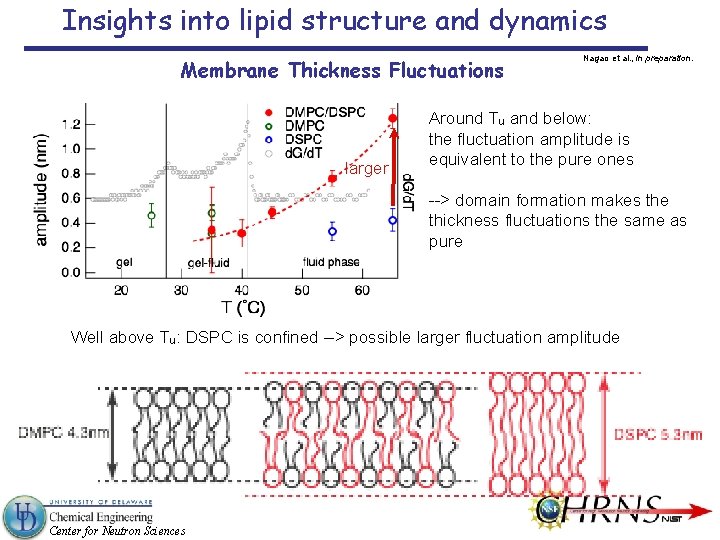
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Width of Lorentz peak relates to the fluctuation amplitude Mean amplitude = 3. 7 Å ± 0. 7 Å experiment Huang’s mean amplitude ≈ 4. 5 Å Huang, Biophys. J. 50, 1061 (1986). theory Lindahl & Edholm’s amplitude ≈ 5 Å Lindahl and Edholm, Biophys. J. 79, 426 (2000). simulation ≈ 8 % of the membrane thickness; close to the value seen in surfactant membranes (≈ 12 %) Is amplitude defined by geometrical constraints, like volume conservation? Center for Neutron Sciences

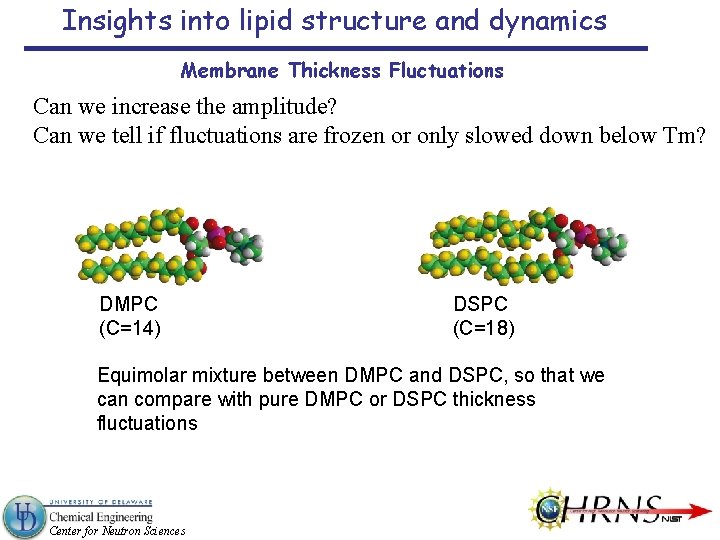
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Can we increase the amplitude? Can we tell if fluctuations are frozen or only slowed down below Tm? DMPC (C=14) DSPC (C=18) Equimolar mixture between DMPC and DSPC, so that we can compare with pure DMPC or DSPC thickness fluctuations Center for Neutron Sciences

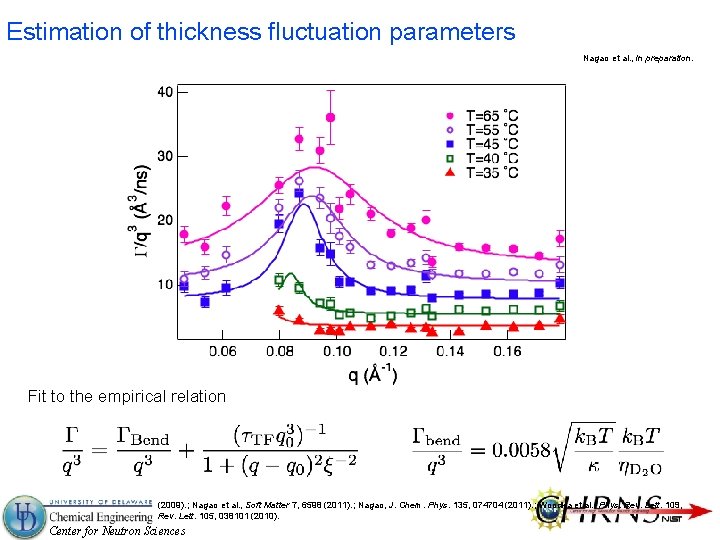
Estimation of thickness fluctuation parameters Nagao et al. , in preparation. Fit to the empirical relation Nagao, Phys. Rev. E 80, 031606 (2009). ; Nagao et al. , Soft Matter 7, 6598 (2011). ; Nagao, J. Chem. Phys. 135, 074704 (2011). ; Woodka et al. , Phys. Rev. Lett. 109, 058102 (2012). ; Lee et al. , Phys. Rev. Lett. 105, 038101 (2010). Center for Neutron Sciences

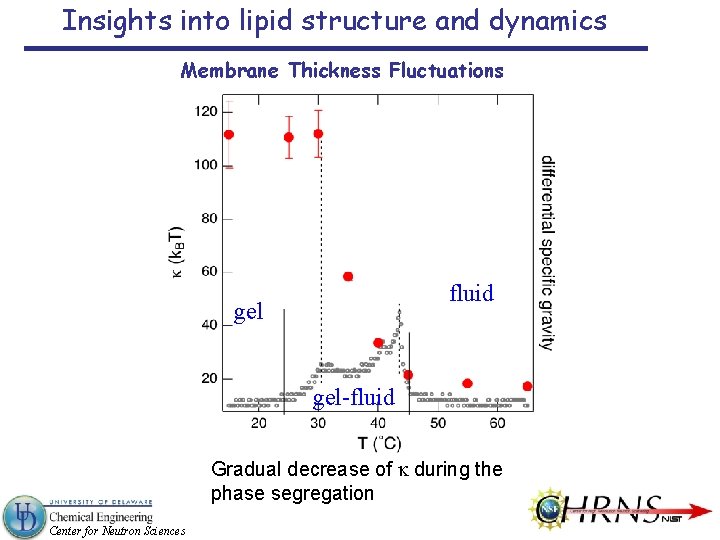
Insights into lipid structure and dynamics Membrane Thickness Fluctuations fluid gel-fluid Gradual decrease of k during the phase segregation Center for Neutron Sciences

Insights into lipid structure and dynamics Membrane Thickness Fluctuations slow down Fluid phase the time scale is similar to the pure cases Gel-Fluid coexistence significantly slower than in the fluid phase Center for Neutron Sciences

Insights into lipid structure and dynamics Membrane Thickness Fluctuations larger Nagao et al. , in preparation. Around Tu and below: the fluctuation amplitude is equivalent to the pure ones --> domain formation makes the thickness fluctuations the same as pure Well above Tu: DSPC is confined --> possible larger fluctuation amplitude Center for Neutron Sciences

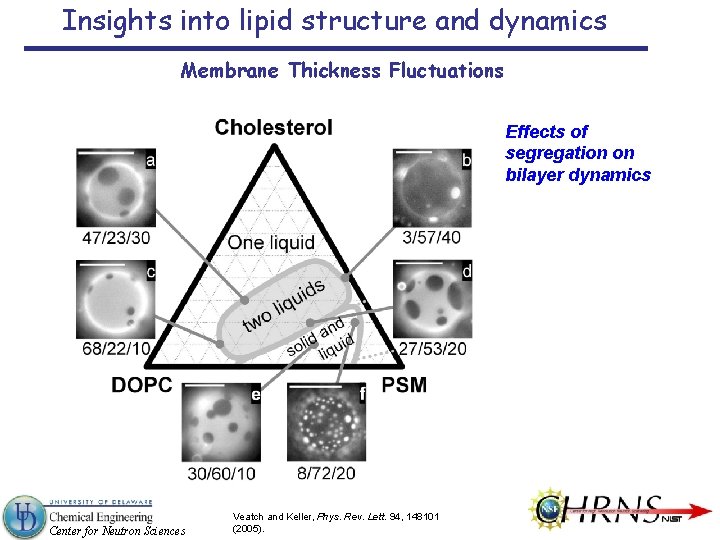
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Effects of segregation on bilayer dynamics Center for Neutron Sciences Veatch and Keller, Phys. Rev. Lett. 94, 148101 (2005).

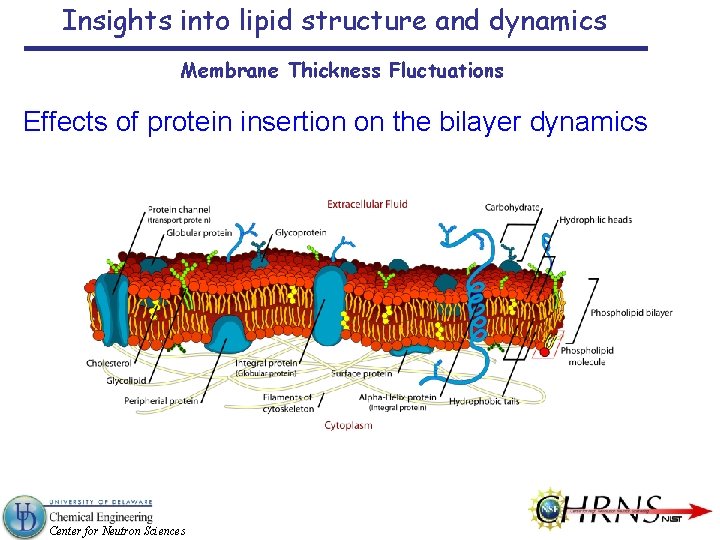
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Effects of protein insertion on the bilayer dynamics Center for Neutron Sciences

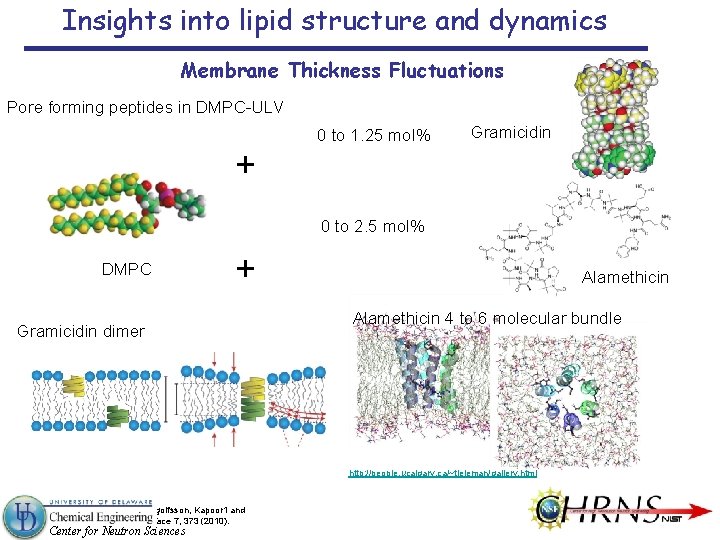
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Pore forming peptides in DMPC-ULV 0 to 1. 25 mol% Gramicidin + 0 to 2. 5 mol% DMPC + Gramicidin dimer Alamethicin 4 to 6 molecular bundle http: //people. ucalgary. ca/~tieleman/gallery. html Lundbæk, Collingwood, Ingolfsson, Kapoor 1 and Andersen, J. R. Soc. Interface 7, 373 (2010). Center for Neutron Sciences

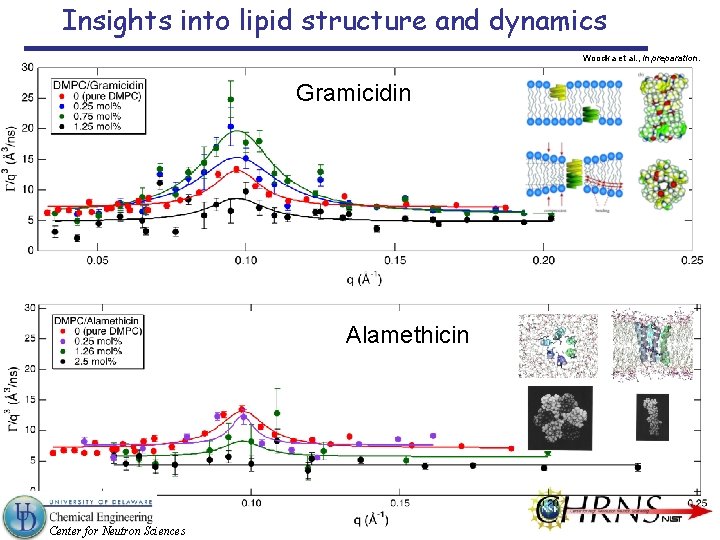
Insights into lipid structure and dynamics Woodka et al. , in preparation. Gramicidin Alamethicin Center for Neutron Sciences

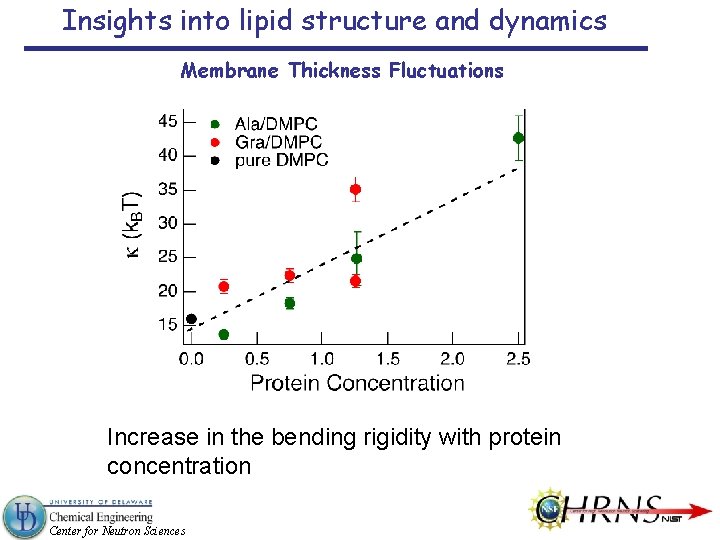
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Increase in the bending rigidity with protein concentration Center for Neutron Sciences

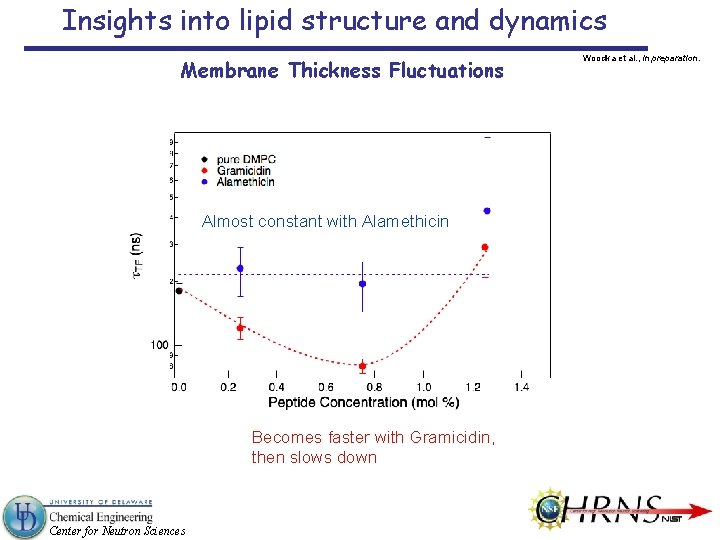
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Almost constant with Alamethicin Becomes faster with Gramicidin, then slows down Center for Neutron Sciences Woodka et al. , in preparation.

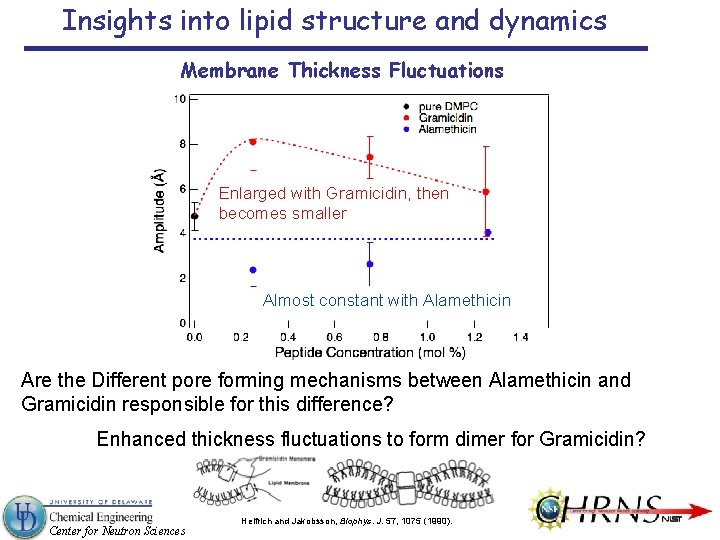
Insights into lipid structure and dynamics Membrane Thickness Fluctuations Enlarged with Gramicidin, then becomes smaller Almost constant with Alamethicin Are the Different pore forming mechanisms between Alamethicin and Gramicidin responsible for this difference? Enhanced thickness fluctuations to form dimer for Gramicidin? Center for Neutron Sciences Helfrich and Jakobsson, Biophys. J. 57, 1075 (1990).

Insights into lipid structure and dynamics Concluding Remarks • Neutrons can be a very powerful tool if used correctly • NS can help answer questions of structure and dynamics over a very wide range of length and time scales • The more work you put into designing the experiment the higher the reward • We have lots of fun!!! • …. . Neutrons Don’t Lie Center for Neutron Sciences