INGID 04 10 2012 Development in genetics and

![Mutations result from mistakes during DNA replication, these events are frequent : nucleotide[s] substitution, Mutations result from mistakes during DNA replication, these events are frequent : nucleotide[s] substitution,](https://slidetodoc.com/presentation_image_h/312cc102c5d5b490663d7802b645f51f/image-5.jpg)
- Slides: 44

INGID 04 -10 -2012 Development in genetics and PID

PIDs • How many PIDs ? A need to identify them, to understand pathophysiology • Diagnosis, treatment, prevention • Most PIDs are inherited monogenic diseases

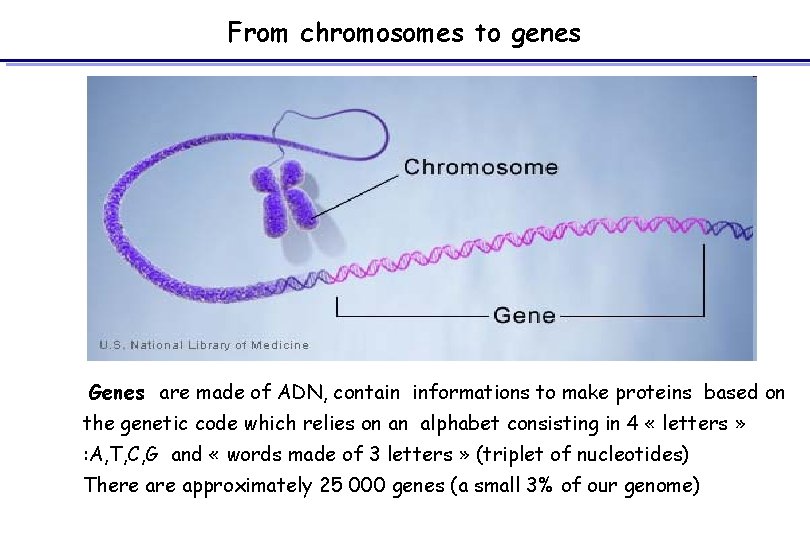
From chromosomes to genes Genes are made of ADN, contain informations to make proteins based on the genetic code which relies on an alphabet consisting in 4 « letters » : A, T, C, G and « words made of 3 letters » (triplet of nucleotides) There approximately 25 000 genes (a small 3% of our genome)

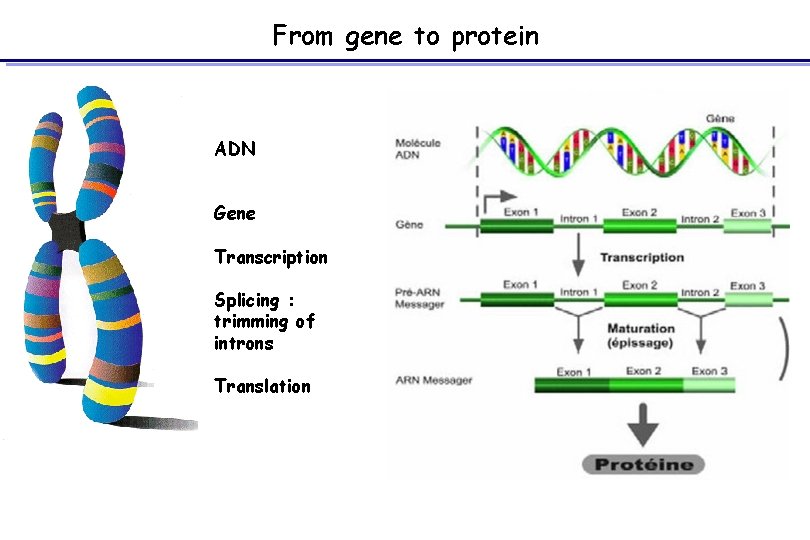
From gene to protein ADN Gene Transcription Splicing : trimming of introns Translation
![Mutations result from mistakes during DNA replication these events are frequent nucleotides substitution Mutations result from mistakes during DNA replication, these events are frequent : nucleotide[s] substitution,](https://slidetodoc.com/presentation_image_h/312cc102c5d5b490663d7802b645f51f/image-5.jpg)
Mutations result from mistakes during DNA replication, these events are frequent : nucleotide[s] substitution, deletion, inversion the new DNA will thus differ from the original copy Most are neutral, some are deleterious

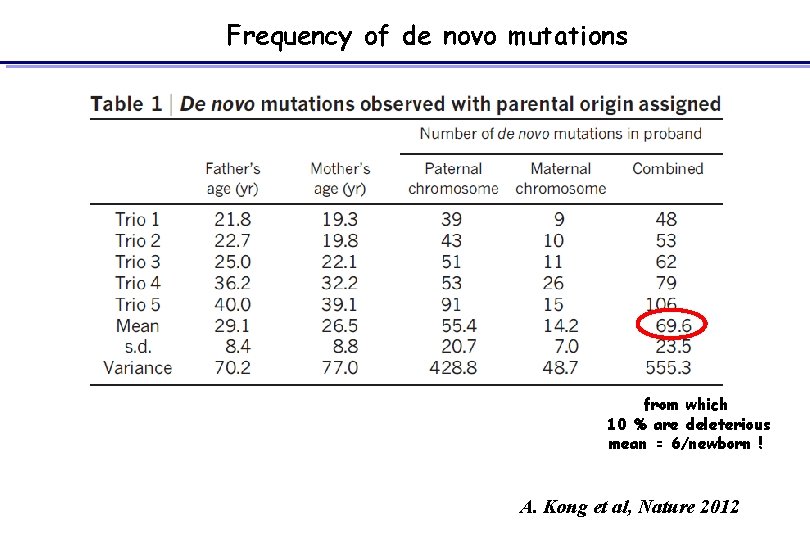
Frequency of de novo mutations from which 10 % are deleterious mean = 6/newborn ! A. Kong et al, Nature 2012

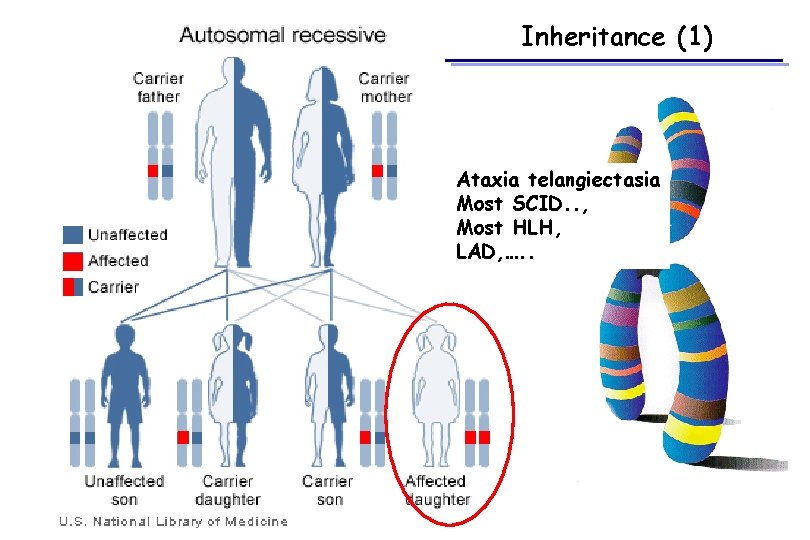
Inheritance (1) Ataxia telangiectasia Most SCID. . , Most HLH, LAD, …. .

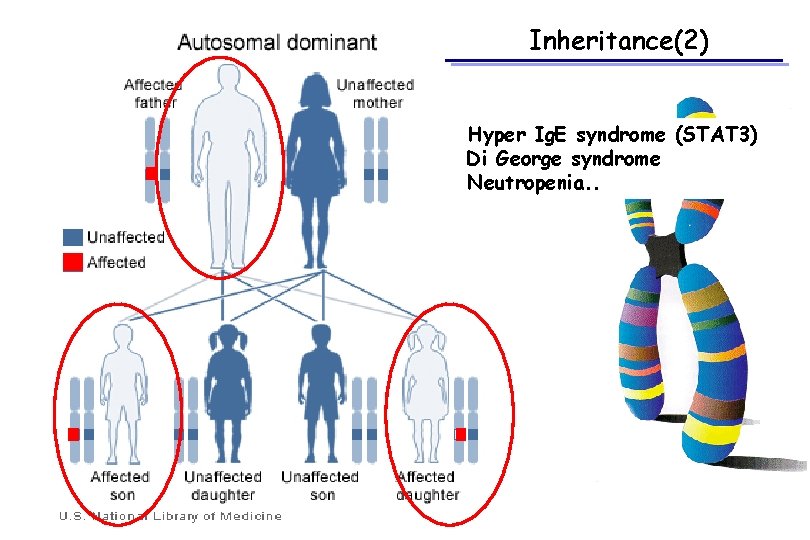
Inheritance(2) Hyper Ig. E syndrome (STAT 3) Di George syndrome Neutropenia. .

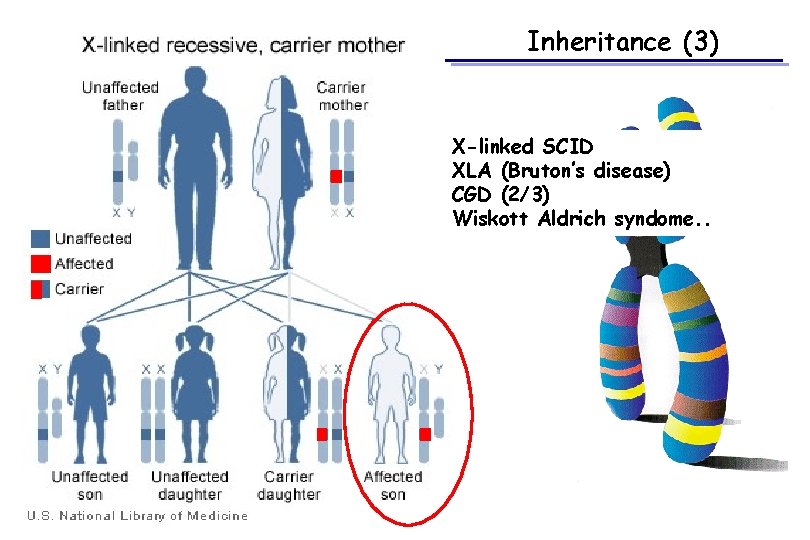
Inheritance (3) X-linked SCID XLA (Bruton’s disease) CGD (2/3) Wiskott Aldrich syndome. .

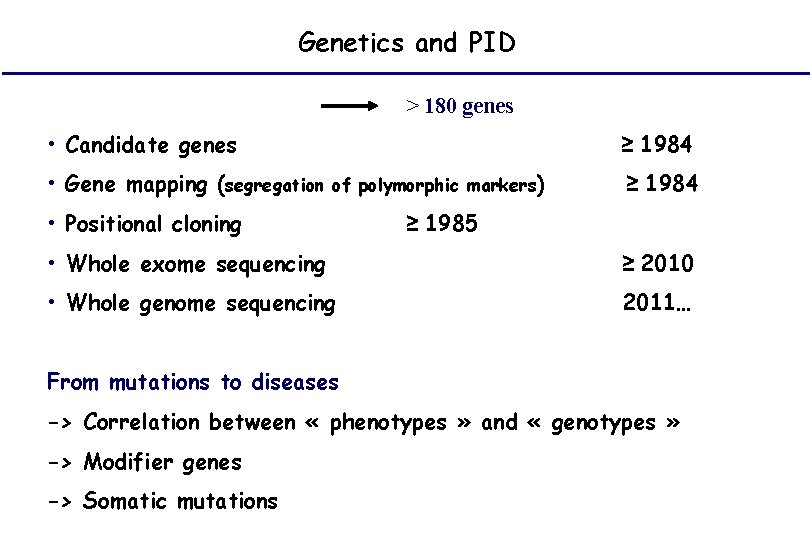
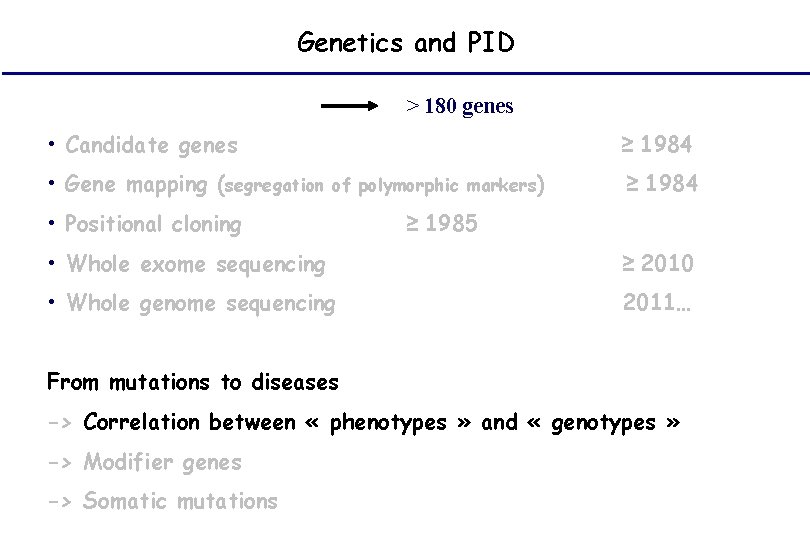
Genetics and PID > 180 genes • Candidate genes • Gene mapping (segregation ≥ 1984 of polymorphic markers) • Positional cloning ≥ 1984 ≥ 1985 • Whole exome sequencing ≥ 2010 • Whole genome sequencing 2011… From mutations to diseases -> Correlation between « phenotypes » and « genotypes » -> Modifier genes -> Somatic mutations

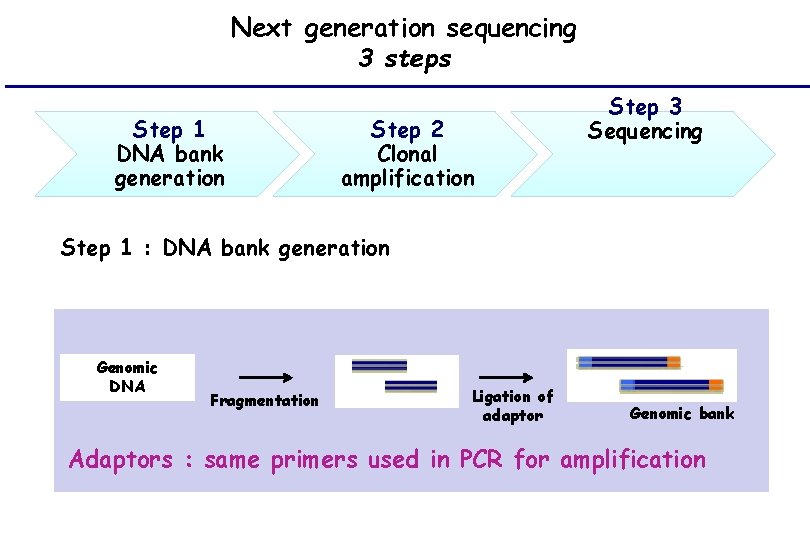
Next generation sequencing 3 steps Step 1 DNA bank generation Step 2 Clonal amplification Step 3 Sequencing Step 1 : DNA bank generation Genomic DNA Fragmentation Ligation of adaptor Genomic bank Adaptors : same primers used in PCR for amplification

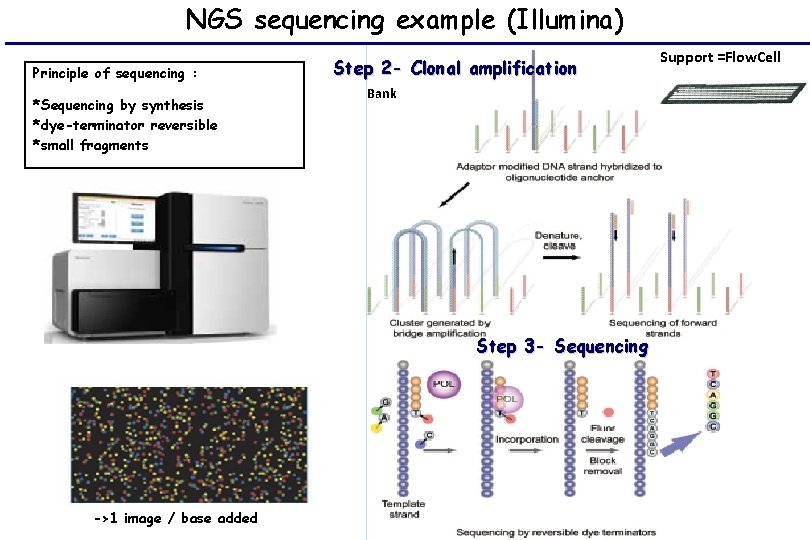
NGS sequencing example (Illumina) Principle of sequencing : *Sequencing by synthesis *dye-terminator reversible *small fragments Step 2 - Clonal amplification Bank Step 3 - Sequencing ->1 image / base added Support =Flow. Cell

Genetics and PID Diagnosis Prognosis Treatement Screening Genetic counseling

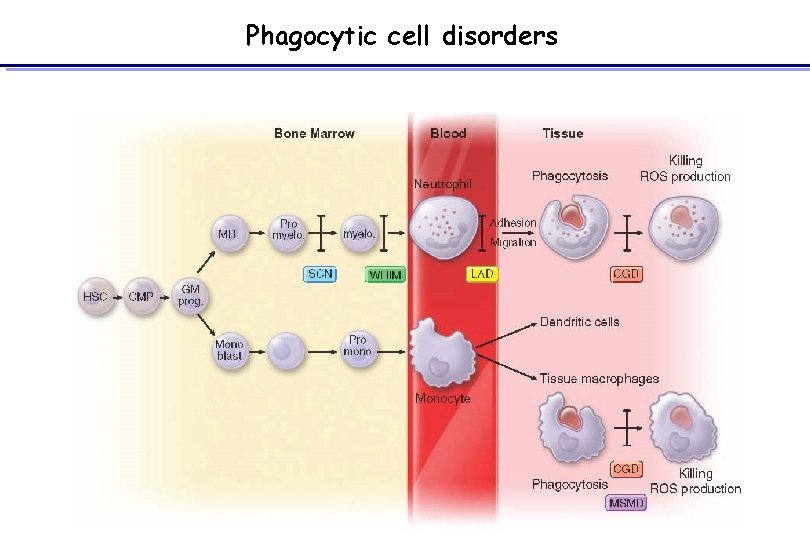
Phagocytic cell disorders

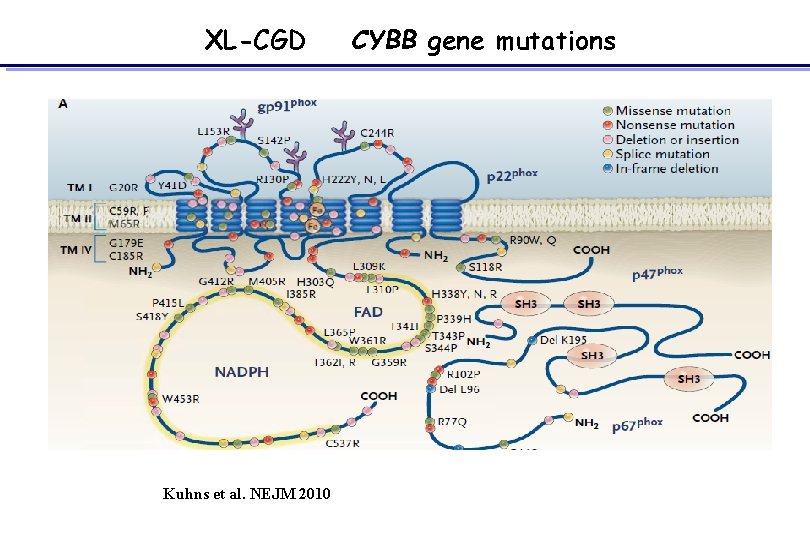
XL-CGD Kuhns et al. NEJM 2010 CYBB gene mutations

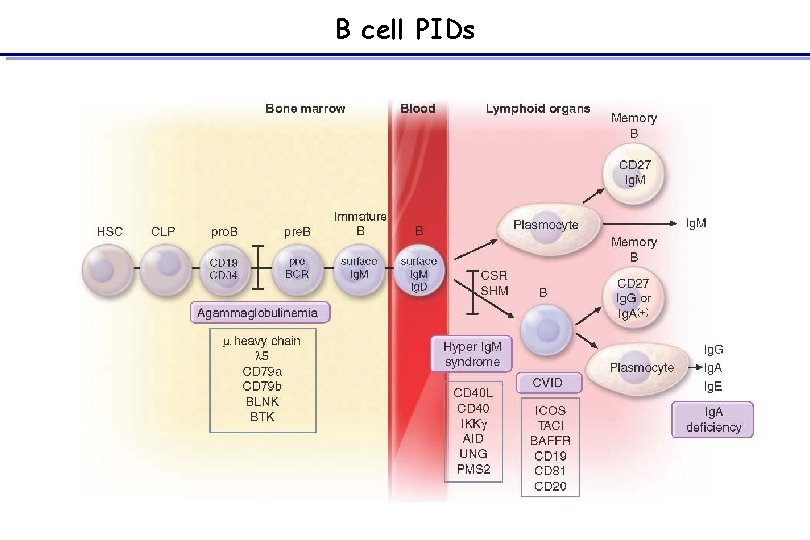
B cell PIDs

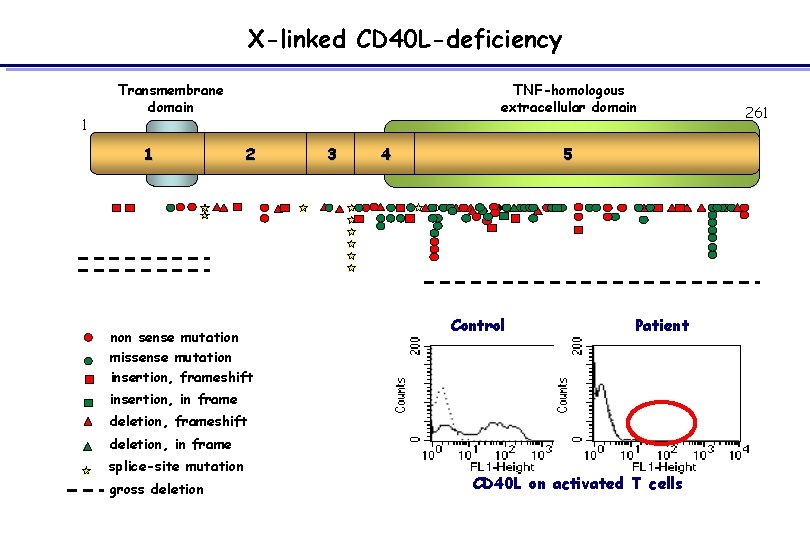
X-linked CD 40 L-deficiency 1 Transmembrane domain 1 TNF-homologous extracellular domain 2 non sense mutation missense mutation insertion, frameshift 3 4 5 Control Patient insertion, in frame deletion, frameshift deletion, in frame splice-site mutation gross deletion CD 40 L on activated T cells 261

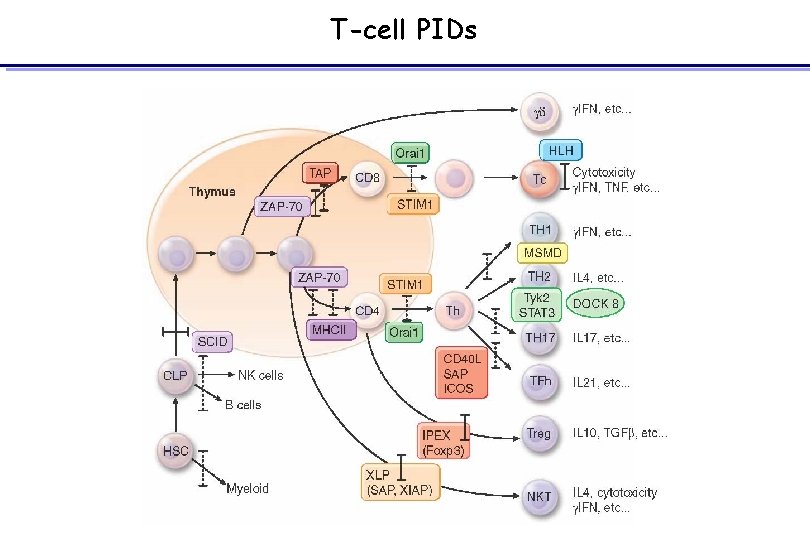
T-cell PIDs

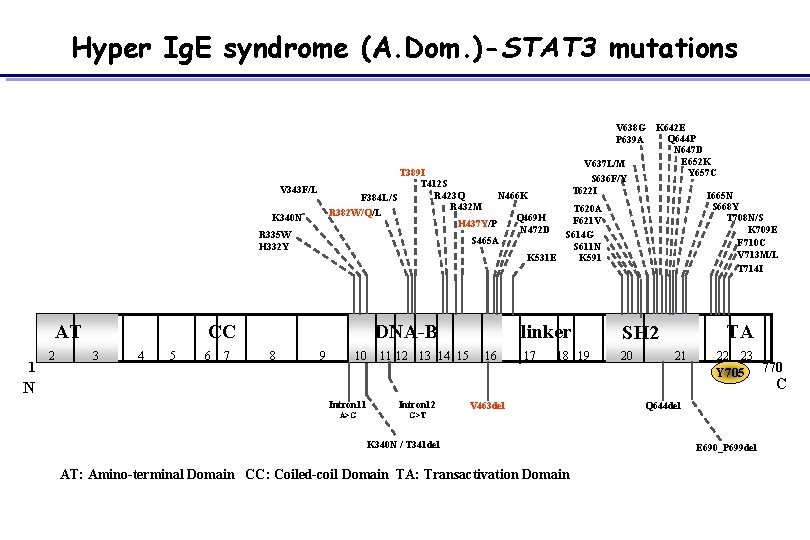
Hyper Ig. E syndrome (A. Dom. )-STAT 3 mutations T 389 I T 412 S R 423 Q N 466 K F 384 L/S R 432 M R 382 W/Q/L Q 469 H H 437 Y/P N 472 D S 465 A V 343 F/L R 335 W H 332 Y 1 2 CC 3 4 5 6 7 DNA-B 8 S 668 Y T 708 N/S K 709 E F 710 C V 713 M/L T 714 I T 620 A F 621 V S 614 G S 611 N K 531 E K 591 K 340 N AT V 638 G K 642 E Q 644 P P 639 A N 647 D E 652 K V 637 L/M Y 657 C S 636 F/Y T 622 I I 665 N 9 10 11 12 13 14 15 16 linker SH 2 17 20 18 19 TA 21 22 23 Y 705 N Intron 11 Intron 12 A>G G>T V 463 del K 340 N / T 341 del AT: Amino-terminal Domain CC: Coiled-coil Domain TA: Transactivation Domain Q 644 del E 690_P 699 del 770 C

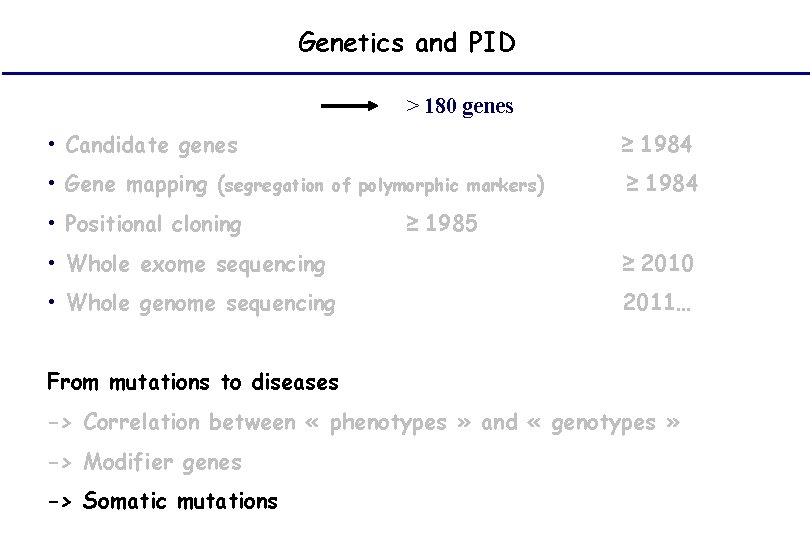
Genetics and PID > 180 genes • Candidate genes • Gene mapping (segregation ≥ 1984 of polymorphic markers) • Positional cloning ≥ 1984 ≥ 1985 • Whole exome sequencing ≥ 2010 • Whole genome sequencing 2011… From mutations to diseases -> Correlation between « phenotypes » and « genotypes » -> Modifier genes -> Somatic mutations

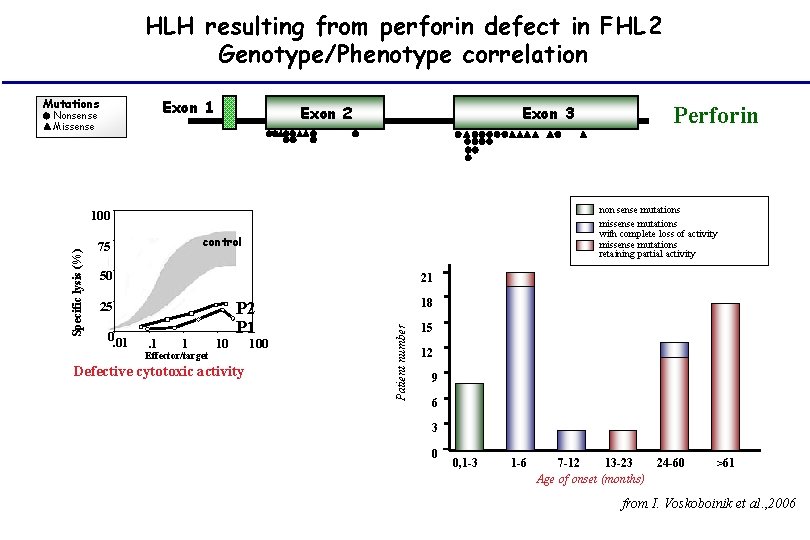
HLH resulting from perforin defect in FHL 2 Genotype/Phenotype correlation Exon 1 Mutations Nonsense Missense Exon 2 Exon 3 non sense mutations missense mutations with complete loss of activity missense mutations retaining partial activity control 50 21 25 0. 01 . 1 1 Effector/target 10 P 2 P 1 Defective cytotoxic activity 100 18 Patient number Specific lysis (%) 100 75 Perforin 15 12 9 6 3 0 0, 1 -3 1 -6 7 -12 13 -23 24 -60 Age of onset (months) >61 from I. Voskoboinik et al. , 2006

Genetics and PID > 180 genes • Candidate genes • Gene mapping (segregation ≥ 1984 of polymorphic markers) • Positional cloning ≥ 1984 ≥ 1985 • Whole exome sequencing ≥ 2010 • Whole genome sequencing 2011… From mutations to diseases -> Correlation between « phenotypes » and « genotypes » -> Modifier genes -> Somatic mutations

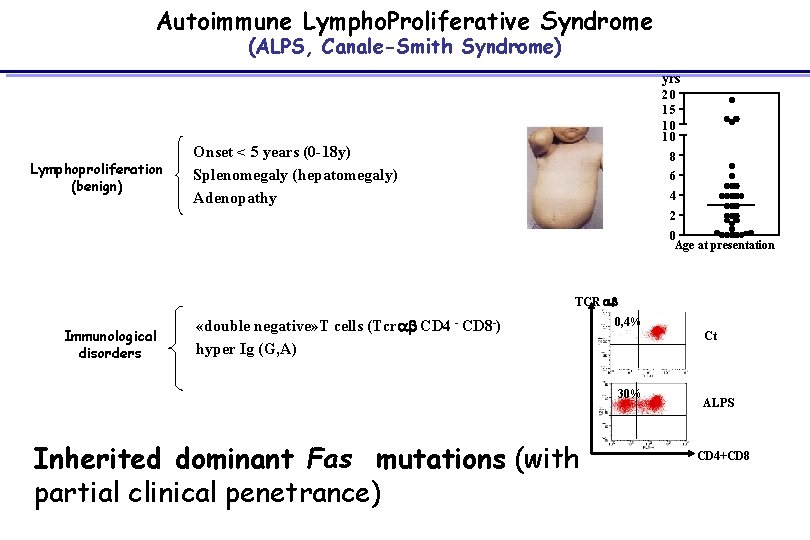
Autoimmune Lympho. Proliferative Syndrome (ALPS, Canale-Smith Syndrome) yrs Lymphoproliferation (benign) 20 15 10 10 Onset < 5 years (0 -18 y) Splenomegaly (hepatomegaly) Adenopathy 8 6 4 2 0 Age at presentation TCR ab Immunological disorders «double negative» T cells (Tcrab CD 4 - CD 8 -) hyper Ig (G, A) autoimmunity (2/3 patients) 0, 4% Ct 30% Inherited dominant Fas mutations (with partial clinical penetrance) ALPS CD 4+CD 8

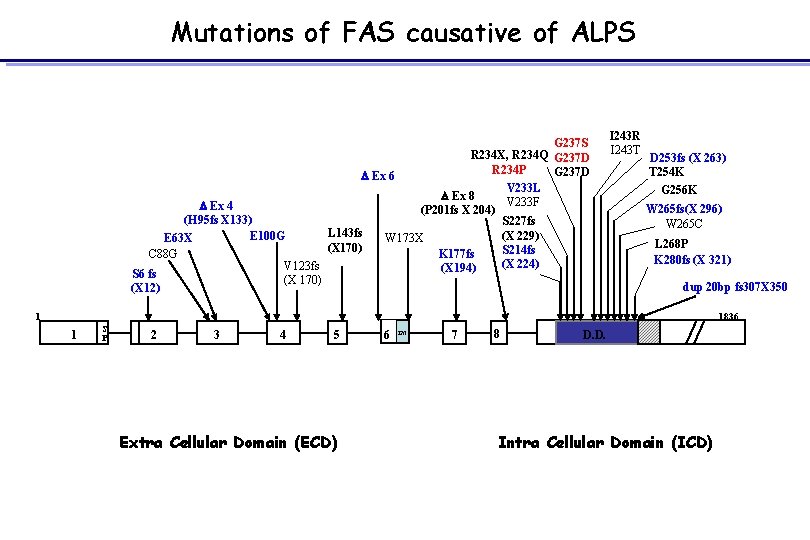
Mutations of FAS causative of ALPS G 237 S R 234 X, R 234 Q G 237 D R 234 P G 237 D D Ex 6 V 233 L D Ex 8 V 233 F D Ex 4 (P 201 fs X 204) (H 95 fs X 133) S 227 fs L 143 fs E 100 G (X 229) W 173 X E 63 X (X 170) S 214 fs C 88 G K 177 fs (X 224) V 123 fs (X 194) S 6 fs (X 170) (X 12) I 243 R I 243 T D 253 fs (X 263) T 254 K G 256 K W 265 fs(X 296) W 265 C L 268 P K 280 fs (X 321) dup 20 bp fs 307 X 350 1836 1 1 S P 2 3 4 5 Extra Cellular Domain (ECD) 6 TM 7 8 D. D. Intra Cellular Domain (ICD)

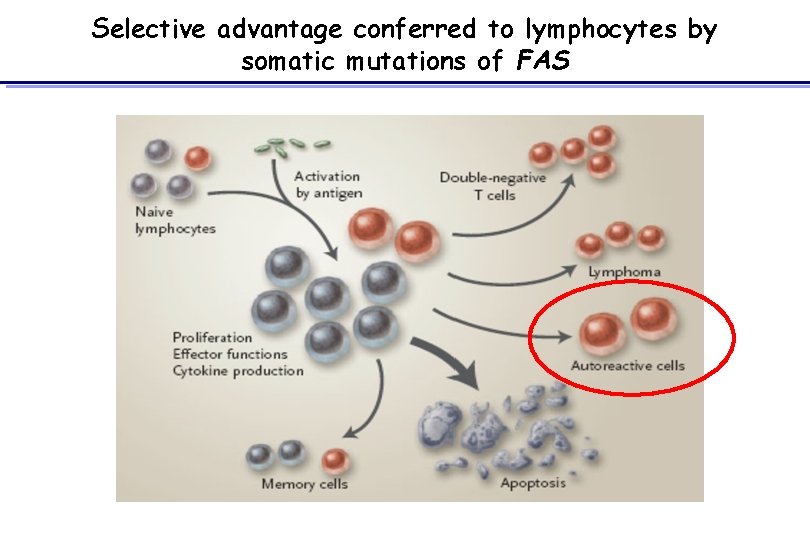
Selective advantage conferred to lymphocytes by somatic mutations of FAS

Genetics and PID • Diagnosis • Prognosis • Treatement • Screening • Genetic counselling

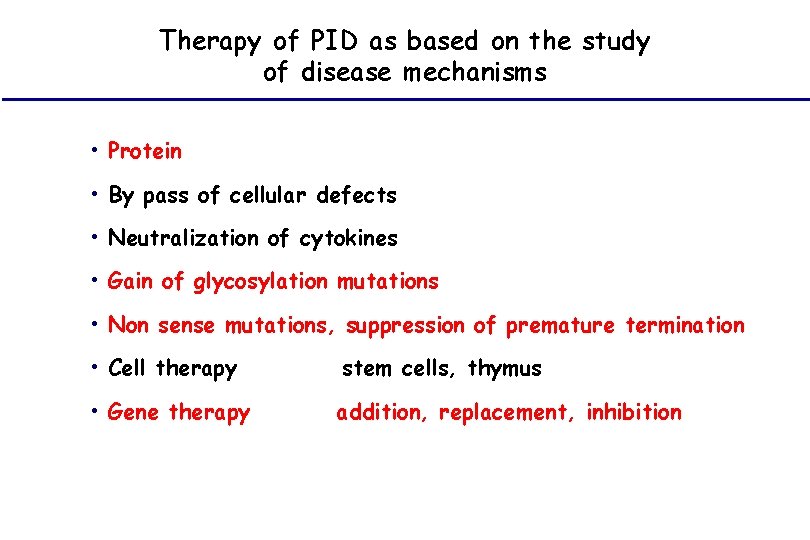
Therapy of PID as based on the study of disease mechanisms • Protein • By pass of cellular defects • Neutralization of cytokines • Gain of glycosylation mutations • Non sense mutations, suppression of premature termination • Cell therapy stem cells, thymus • Gene therapy addition, replacement, inhibition

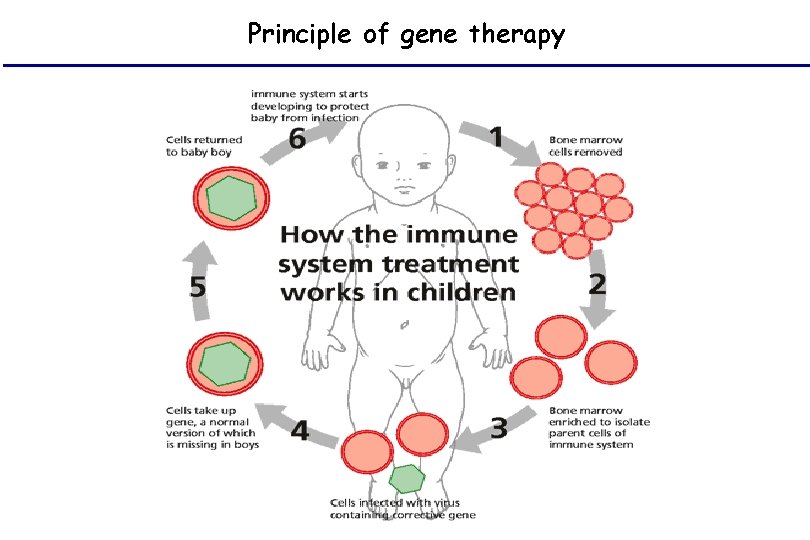
Principle of gene therapy

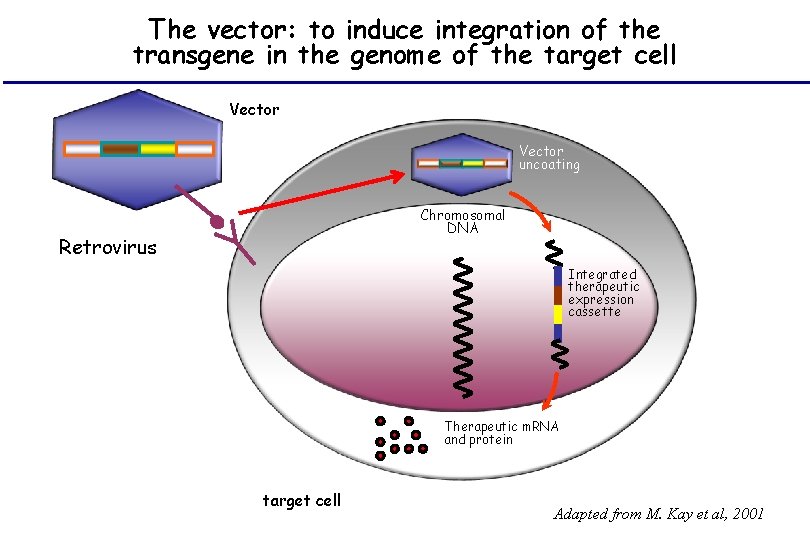
The vector: to induce integration of the transgene in the genome of the target cell Vector uncoating Chromosomal DNA Retrovirus Integrated therapeutic expression cassette Therapeutic m. RNA and protein target cell Adapted from M. Kay et al, 2001

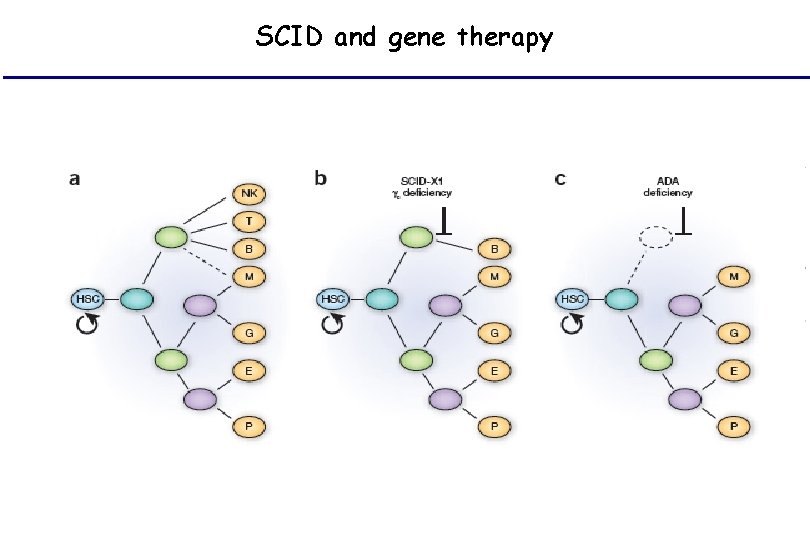
SCID and gene therapy

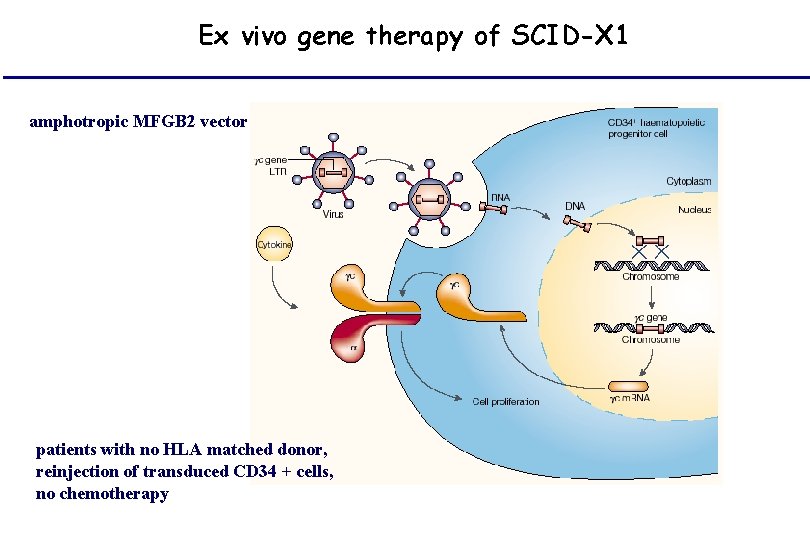
Ex vivo gene therapy of SCID-X 1 amphotropic MFGB 2 vector patients with no HLA matched donor, reinjection of transduced CD 34 + cells, no chemotherapy

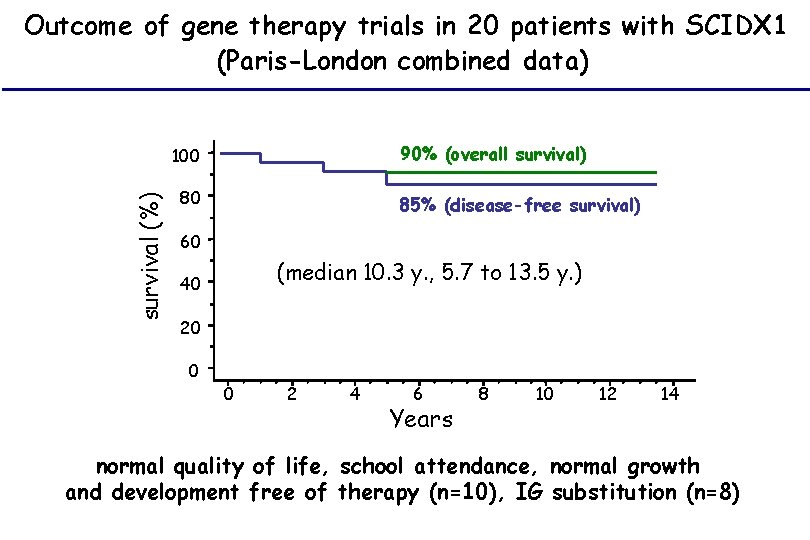
Outcome of gene therapy trials in 20 patients with SCIDX 1 (Paris-London combined data) 90% (overall survival) survival (%) 100 80 85% (disease-free survival) 60 (median 10. 3 y. , 5. 7 to 13. 5 y. ) 40 20 0 0 2 4 6 Years 8 10 12 14 normal quality of life, school attendance, normal growth and development free of therapy (n=10), IG substitution (n=8)

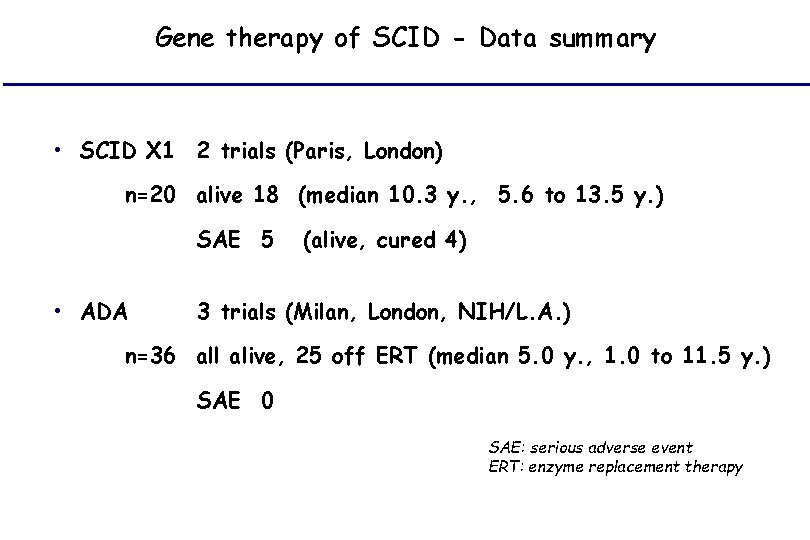
Gene therapy of SCID - Data summary • SCID X 1 2 trials (Paris, London) n=20 alive 18 (median 10. 3 y. , 5. 6 to 13. 5 y. ) SAE 5 • ADA (alive, cured 4) 3 trials (Milan, London, NIH/L. A. ) n=36 all alive, 25 off ERT (median 5. 0 y. , 1. 0 to 11. 5 y. ) SAE 0 SAE: serious adverse event ERT: enzyme replacement therapy

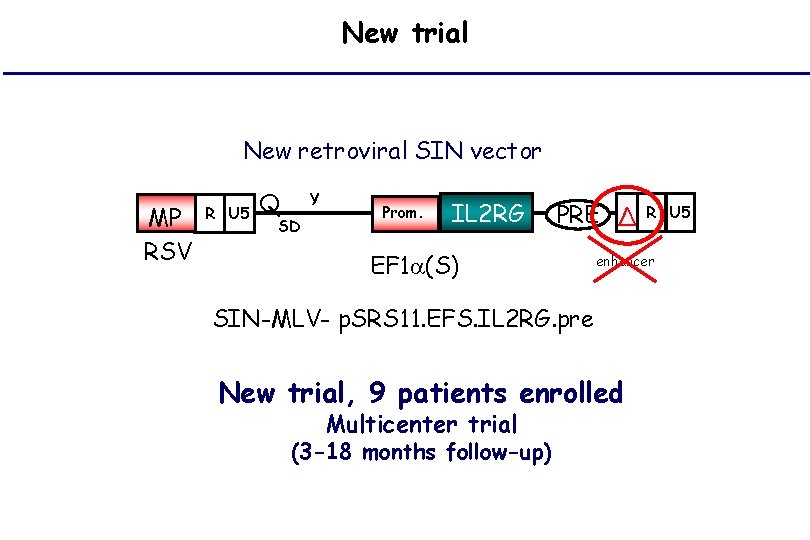
New trial New retroviral SIN vector MP RSV R U 5 Q Y SD Prom. IL 2 RG PRE Δ EF 1 (S) enhancer SIN-MLV- p. SRS 11. EFS. IL 2 RG. pre New trial, 9 patients enrolled Multicenter trial (3 -18 months follow-up) R U 5

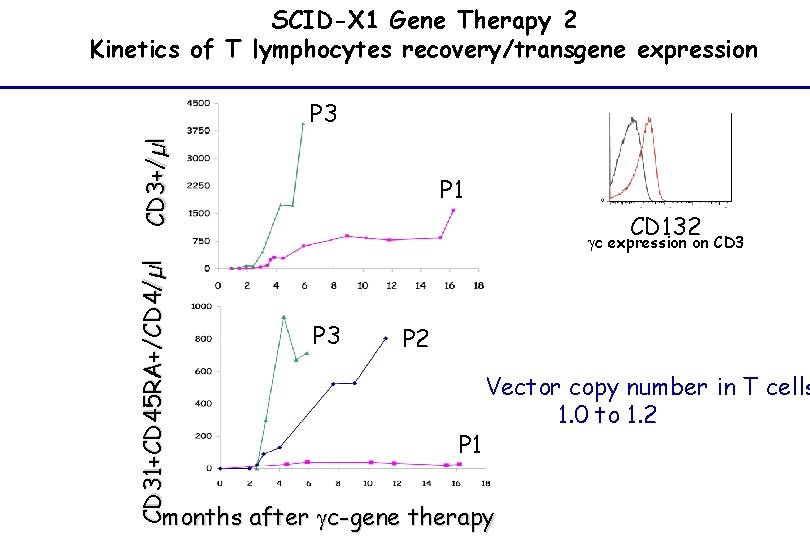
SCID-X 1 Gene Therapy 2 Kinetics of T lymphocytes recovery/transgene expression CD 3+/µl P 3 P 1 CD 132 CD 31+CD 45 RA+/CD 4/µl gc expression on CD 3 P 2 P 1 Vector copy number in T cells 1. 0 to 1. 2 months after gc-gene therapy

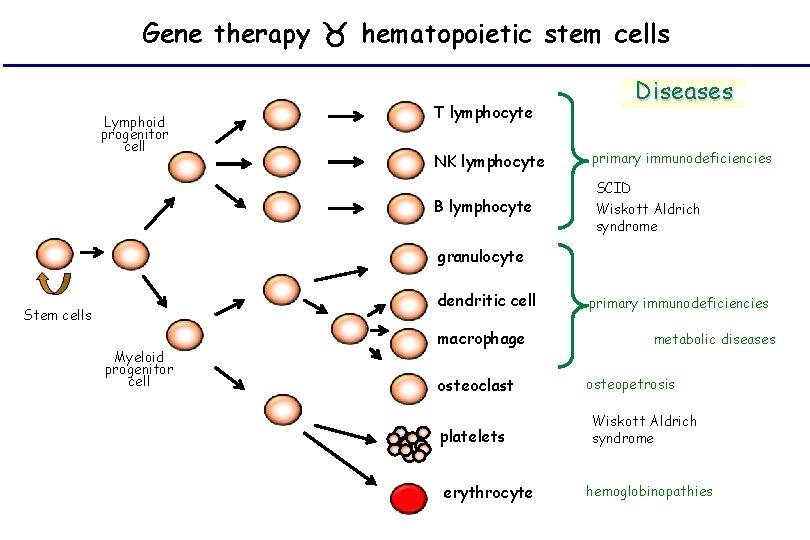
Gene therapy hematopoietic stem cells Lymphoid progenitor cell T lymphocyte Diseases NK lymphocyte primary immunodeficiencies B lymphocyte SCID Wiskott Aldrich syndrome granulocyte dendritic cell Stem cells Myeloid progenitor cell macrophage osteoclast platelets erythrocyte primary immunodeficiencies metabolic diseases osteopetrosis Wiskott Aldrich syndrome hemoglobinopathies

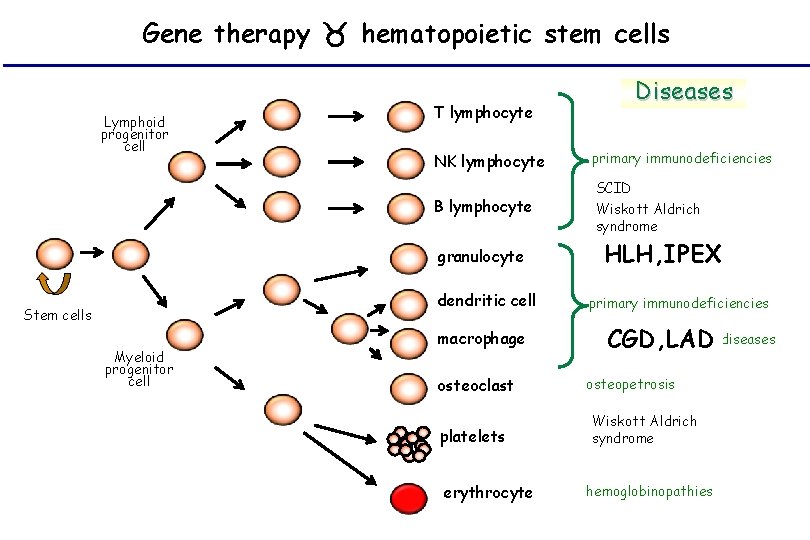
Gene therapy hematopoietic stem cells Lymphoid progenitor cell T lymphocyte NK lymphocyte primary immunodeficiencies B lymphocyte SCID Wiskott Aldrich syndrome granulocyte dendritic cell Stem cells Myeloid progenitor cell Diseases macrophage osteoclast platelets erythrocyte HLH, IPEX primary immunodeficiencies metabolic diseases CGD, LAD osteopetrosis Wiskott Aldrich syndrome hemoglobinopathies

Genetics and PID • Diagnosis • Prognosis • Treatement • Screening • Genetic counseling

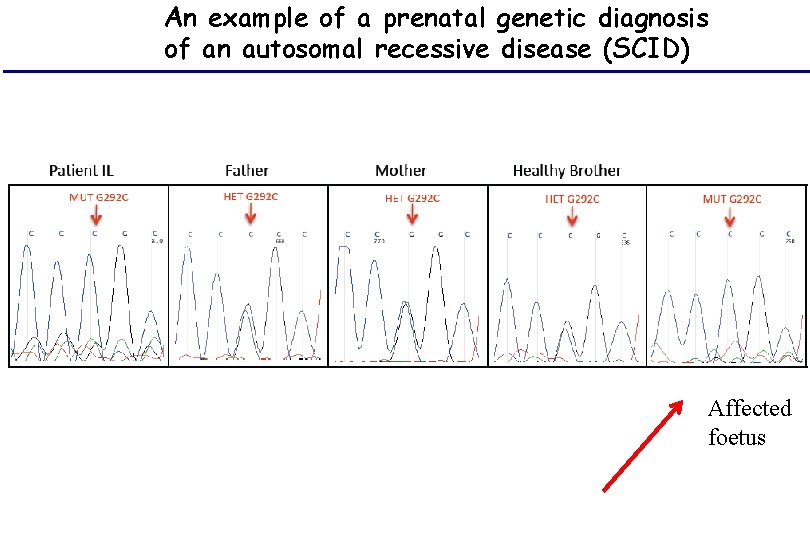
An example of a prenatal genetic diagnosis of an autosomal recessive disease (SCID) Affected foetus

Genetics and PID • Diagnosis • Prognosis • Treatement • Screening • Genetic counselling

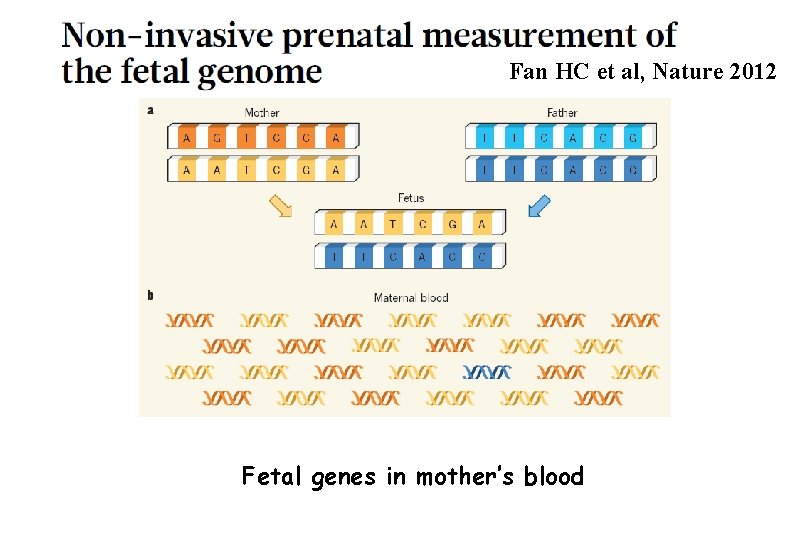
Fan HC et al, Nature 2012 Fetal genes in mother’s blood

Conclusion • Genetic diagnosis will enter more and more frequent daily practice of PID medicine • Monogenic will turn to more complex (genome influence) • Therapeutic development expected • Preimplantatory and Prenatal diagnosis made easier • (Pre)neonatal screening • Ethical issues

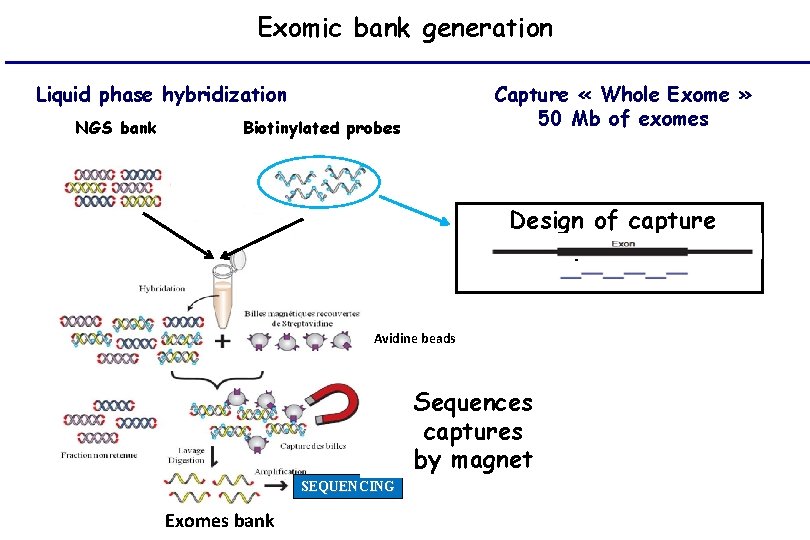
Exomic bank generation Liquid phase hybridization NGS bank Capture « Whole Exome » 50 Mb of exomes Biotinylated probes Design of capture phases Avidine beads Sequences captures by magnet SEQUENCING Exomes bank

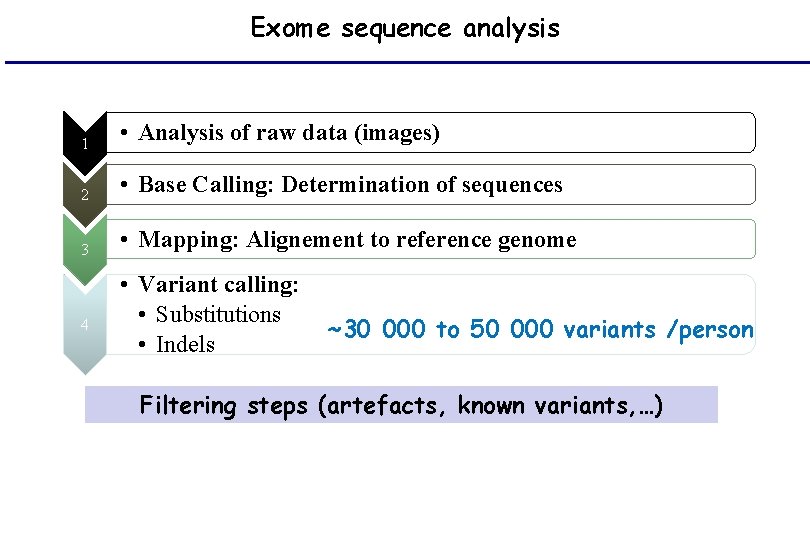
Exome sequence analysis 1 • Analysis of raw data (images) 2 • Base Calling: Determination of sequences 3 • Mapping: Alignement to reference genome 4 • Variant calling: • Substitutions • Indels ~30 000 to 50 000 variants /person Filtering steps (artefacts, known variants, …)