Infrastructure for Automated Data Analysis IADA Antanas Norkus

![GLOBAL CONFIGURATION FILE: AN EXAMPLE [MAIN] # Compulsory parameters run. Prog = root. Gui GLOBAL CONFIGURATION FILE: AN EXAMPLE [MAIN] # Compulsory parameters run. Prog = root. Gui](https://slidetodoc.com/presentation_image/49432ed727d3ac8c9c96eea8c0dad23c/image-7.jpg)





- Slides: 14

Infrastructure for Automated Data Analysis (IADA) Antanas Norkus, Vilnius University, email: antanas. norkus@cern. ch Danilo Piparo, CERN

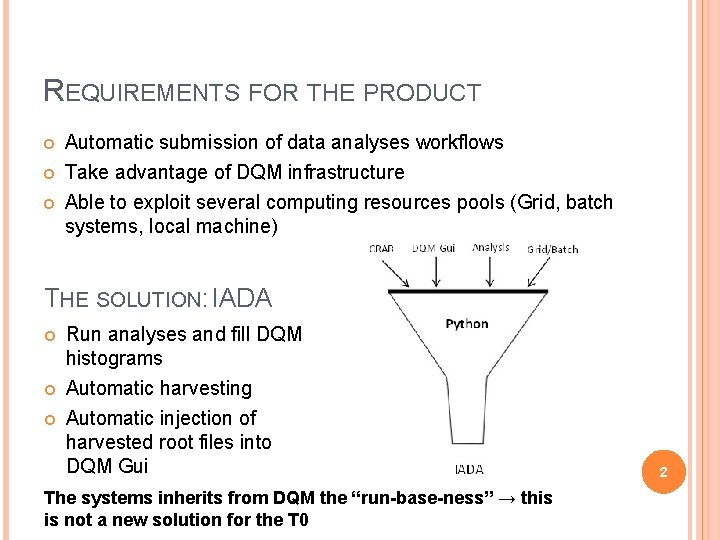
REQUIREMENTS FOR THE PRODUCT Automatic submission of data analyses workflows Take advantage of DQM infrastructure Able to exploit several computing resources pools (Grid, batch systems, local machine) THE SOLUTION: IADA Run analyses and fill DQM histograms Automatic harvesting Automatic injection of harvested root files into DQM Gui The systems inherits from DQM the “run-base-ness” → this is not a new solution for the T 0 2

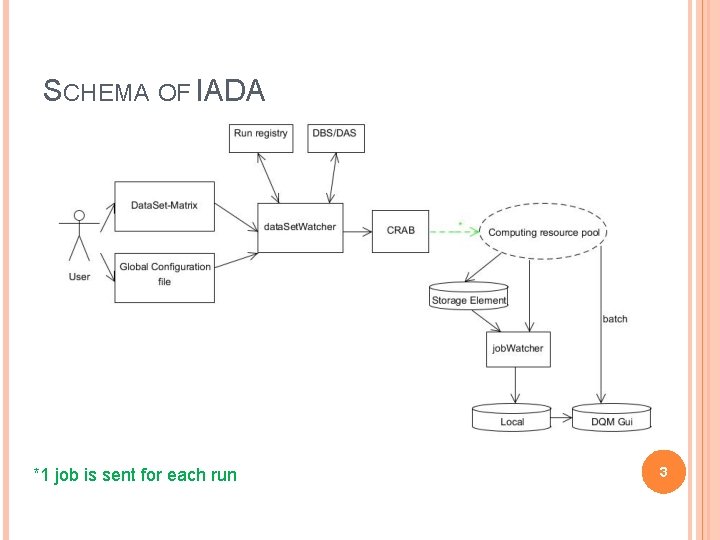
SCHEMA OF IADA *1 job is sent for each run 3

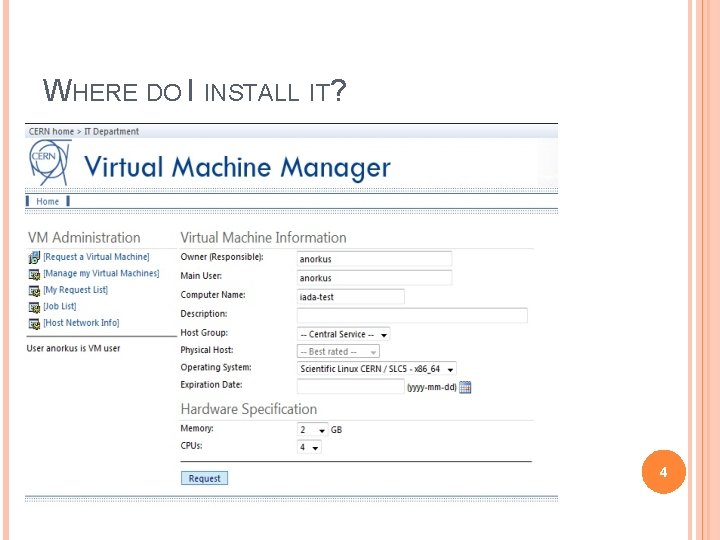
WHERE DO I INSTALL IT? 4

SET-UP COMMANDS Set up CMSSW SCRAM_ARCH=slc 5_amd 64_gcc 462 source /afs/cern. ch/cms/LCG-2/UI/cms_ui_env. sh cmsrel CMMSW_5_2_3_patch 3 cd CMSSW_5_2_3_partch 3/src cmsenv Set up the grid environment and CRAB voms-proxy-init -voms cms myproxy-init -d -n source /afs/cern. ch/cms/ccs/wm/scripts/Crab/crab. sh Setup IADA export IADAWORKDIR=My. IADAWork. Dir svn co https: //svn. cern. ch/reps/Pdm. VTools/IADA/trunk $IADAWORKDIR cd $IADAWORKDIR Setup DQM Gui mkdir DQMGui cd DQMGui svn co svn+ssh: //svn. cern. ch/reps/CMSDMWM/Infrastructure/trunk/Deployment cfg $PWD/cfg/Deploy -R cmsweb@1203 h -t MYDEV -s "prep sw post" $PWD dqmgui/bare $PWD/current/config/dqmgui/manage -f dev start "I did read doc sudo /sbin/iptables -I RH-Firewall-1 -INPUT 3 -p tcp -m tcp --dport 8060 -j ACCEPTumentation" 5

HOW DO I CONFIGURE IT? All the configuration parameters are contained in two places: � Global configuration file: DQM url CRAB Storage Element name E-mail of sender E-mails of recipients � Data. Set. Matrix: Dataset name Set of runs CMSSW python configuration file or cms. Driver command Backend to be used (one of: Grid, caf/lsf, local machine) Json_masking: (3 possibilities ) yes – use RR 3 API to generate lumi_mask file no – use lumi_mask [0: 100000] /path/to/user/predefined/file 6
![GLOBAL CONFIGURATION FILE AN EXAMPLE MAIN Compulsory parameters run Prog root Gui GLOBAL CONFIGURATION FILE: AN EXAMPLE [MAIN] # Compulsory parameters run. Prog = root. Gui](https://slidetodoc.com/presentation_image/49432ed727d3ac8c9c96eea8c0dad23c/image-7.jpg)
GLOBAL CONFIGURATION FILE: AN EXAMPLE [MAIN] # Compulsory parameters run. Prog = root. Gui #the way program works 3 possibilities root. Gui, root. Loc, #root. SE def. Backend = caf #default backend to use if not defined in data. Set. Matrix DQMurl = http: //myserver: 8060 #DQM server address # Optional Parameters def. Queue. Time = 60 #job queue time parameter Email_send_To = mail 1@provider 1, mail 2@provider 2 #addresses recipients of #IADA mails Email_send_From = mail 1@provider 1 # email sender address storage_element = my. SE #name of CRAB storage element. Example: T 2_IT_Legnaro 7

FULL DATASET-MATRIX: AN EXAMPLE {"Process 1": { "dataset": "/Single. Mu/Run 2011 B-Prompt. Reco-v 1/DQM", "config_file": ["/afs/cern. ch/user/a/anorkus/scratch 0/CMSSW_4_2_8/src/DQM/Data. Scouting /test/harvest. py"], "global_tag": "GR_R_42_V 25: : All", "backend": "grid", "min_run": 177548, See next slide "max_run": 177788, "run. Prog": "root. Loc", "queue. Time": 60, “json-filtered: “: “yes" "meta_data": "grid 04 -10 -root. Loc-test"} Will appear in the dataset name in the DQM server } For the cms. Driver usage see the back-up slide 8

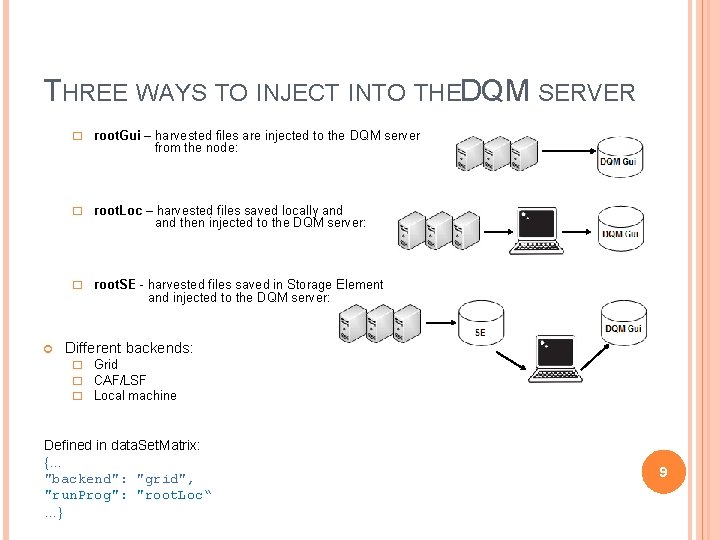
THREE WAYS TO INJECT INTO THEDQM SERVER � root. Gui – harvested files are injected to the DQM server from the node: � root. Loc – harvested files saved locally and then injected to the DQM server: � root. SE - harvested files saved in Storage Element and injected to the DQM server: Different backends: � Grid � CAF/LSF � Local machine Defined in data. Set. Matrix: {… "backend": "grid", "run. Prog": "root. Loc“ as …} 9

SPIN-OFFS Use it for automatic harvesting, including Rel. Val harvesting. � Test your DQM modules. � Possible target: DQM and computing operators Possible target: DQM developers See DQM modules results on already prompt reconstructed data � Possible targets: everybody? 10

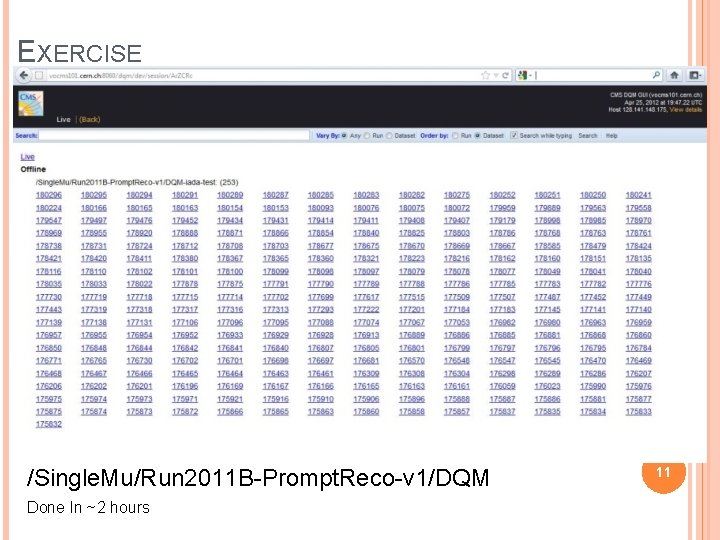
EXERCISE /Single. Mu/Run 2011 B-Prompt. Reco-v 1/DQM Done In ~2 hours 11

POSSIBLE EXTENSIONS Overcome the run-based nature of DQM → “hadd” of harvested files with subsequent injection Auto update of the dataset matrix for adding new datasets � Easy “plug-in” to implement (basically decoupled from existing system) 12

CONCLUSIONS An innovative product was implemented which allows: � Automated analyses (high potential for discovery searches) � Automated harvesting (even on growing datasets) � Test new DQM modules or updates of existing histograms � See outcome of new modules on already processed promptly reconstructed data Twiki documentation: https: //twiki. cern. ch/twiki/bin/viewauth/CMS/Pdm. VIADA 13

BACK-UP cms. Driver command example: cms. Driver. py my. Harvest -s HARVESTING: dqm. Harvesting --conditions @@@THE_GT@@@ --filein file: file 1. root --data --no_exec @@@THE_GT@@@ OR - is changed automatically to Global. Tag defined in data. Set. Matrix ___THE_GT___ Also supports � No change: cms. Driver. py my. Harvest -s HARVESTING: dqm. Harvesting --conditions GR_R_42_V 25: : All --filein file: file 1. root --data --no_exec 14