Increased Crop Yield Through Improved Photosynthesis 4 th



![Example 2: Wheat Leaf CO 2 -responsive Gene Expression Atlas Low [CO 2] Ambient Example 2: Wheat Leaf CO 2 -responsive Gene Expression Atlas Low [CO 2] Ambient](https://slidetodoc.com/presentation_image/5107c0415a18a3ad6316d0f65bad5255/image-16.jpg)


- Slides: 30

Increased Crop Yield Through Improved Photosynthesis 4 th International Conference on Agriculture and Horticulture Beijing, July 14, 2015 Andy Renz, Vice President Business Development

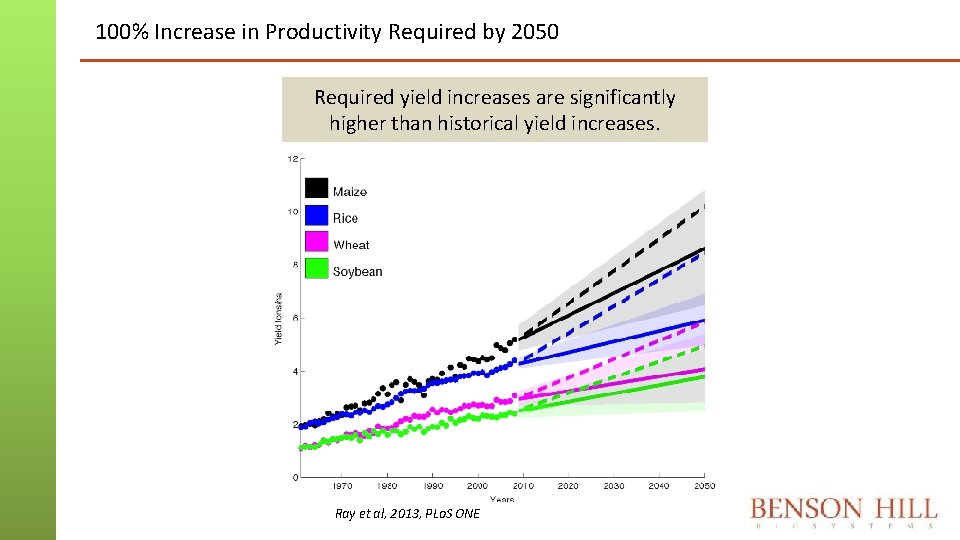
100% Increase in Productivity Required by 2050 Required yield increases are significantly higher than historical yield increases. Ray et al, 2013, PLo. S ONE

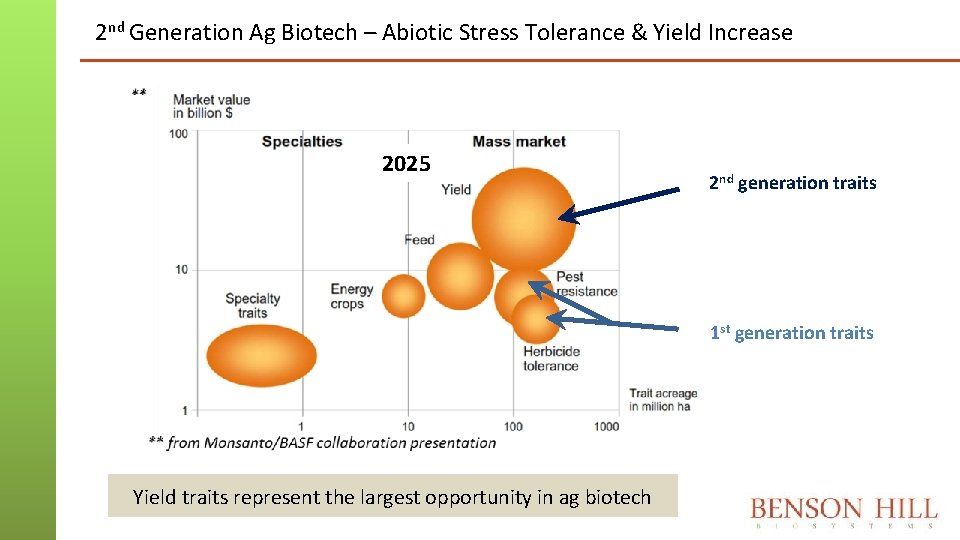
2 nd Generation Ag Biotech – Abiotic Stress Tolerance & Yield Increase 2025 2 nd generation traits 1 st generation traits Yield traits represent the largest opportunity in ag biotech

2 nd Generation Ag Biotech – Abiotic Stress Tolerance & Yield Increase • Excellent results and products from molecular breeding – AQUAmax™ corn from Pioneer – Artesian™ corn from Syngenta – Droght tolerant rice from IRRI • Opportunity and Challenge for GM approaches: – Monsanto/BASF: largest partnership in the history of Ag Biotech R&D: $2. 5 billion(!) • HTP screens in model and crop plats • Field testing in crops (commercial germplasm) • First prducts: Droughtgard™ corn launched in 2013 (Csp. B) – Benson Hill Biosystems: • Focus on yield improvement through improved photosynthesis

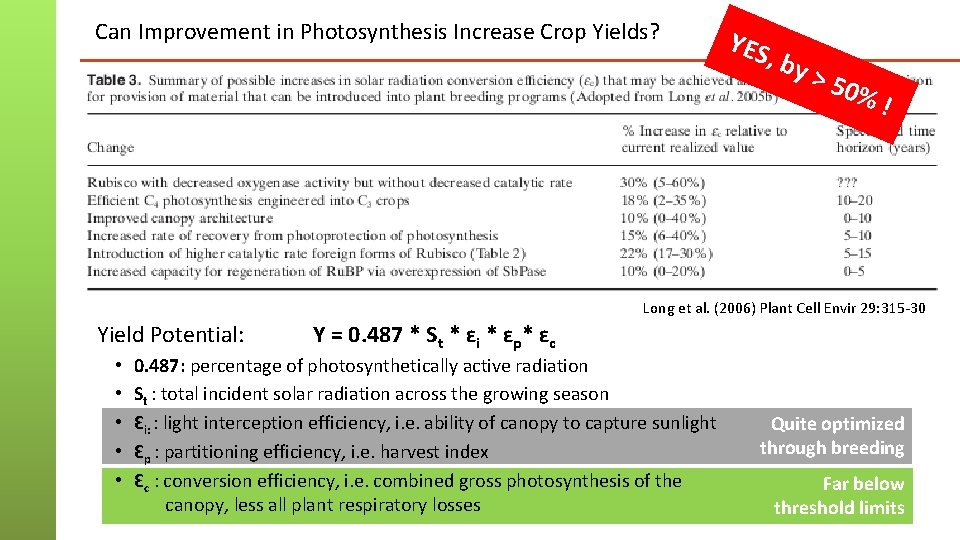
Can Improvement in Photosynthesis Increase Crop Yields? YES , by > 50 %! Long et al. (2006) Plant Cell Envir 29: 315 -30 Yield Potential: • • • Y = 0. 487 * St * ɛi * ɛp* ɛc 0. 487: percentage of photosynthetically active radiation St : total incident solar radiation across the growing season Ɛi: : light interception efficiency, i. e. ability of canopy to capture sunlight Ɛp : partitioning efficiency, i. e. harvest index Ɛc : conversion efficiency, i. e. combined gross photosynthesis of the canopy, less all plant respiratory losses Quite optimized through breeding Far below threshold limits

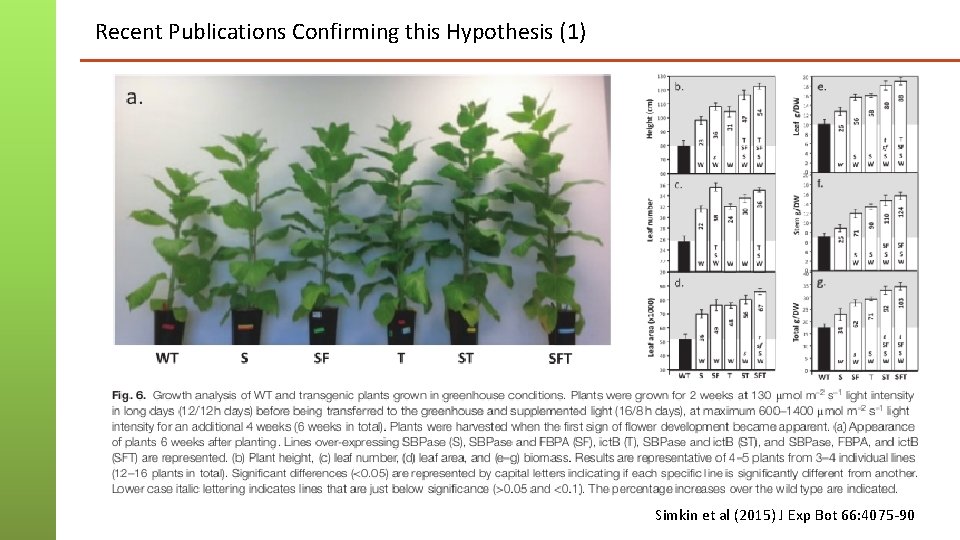
Recent Publications Confirming this Hypothesis (1) Simkin et al (2015) J Exp Bot 66: 4075 -90

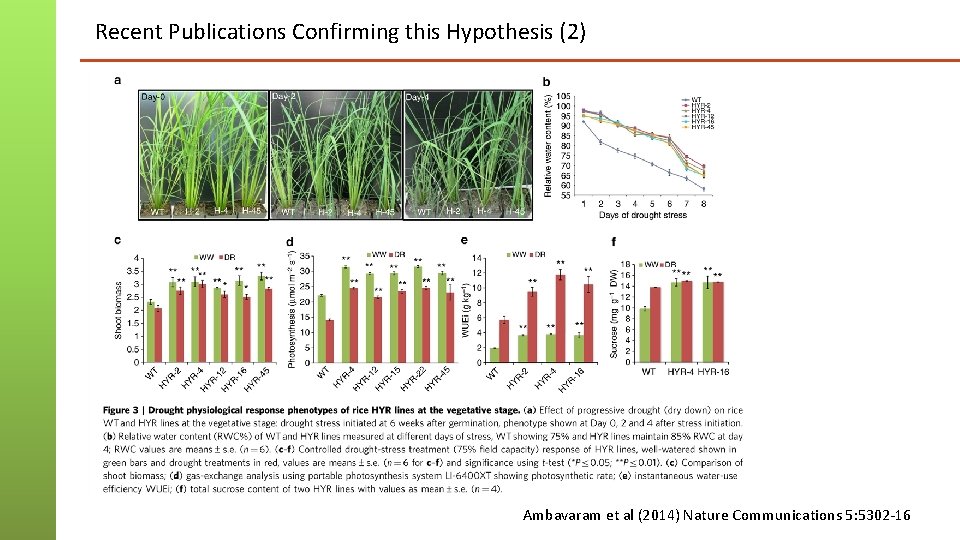
Recent Publications Confirming this Hypothesis (2) Ambavaram et al (2014) Nature Communications 5: 5302 -16

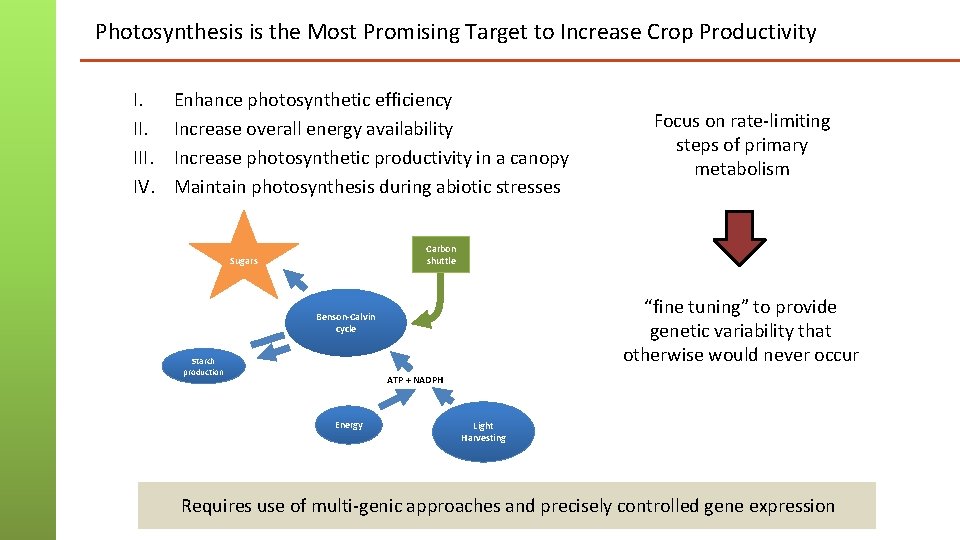
Photosynthesis is the Most Promising Target to Increase Crop Productivity I. III. IV. Enhance photosynthetic efficiency Increase overall energy availability Increase photosynthetic productivity in a canopy Maintain photosynthesis during abiotic stresses Focus on rate-limiting steps of primary metabolism Carbon shuttle Sugars “fine tuning” to provide genetic variability that otherwise would never occur Benson-Calvin cycle Starch production ATP + NADPH Energy Light Harvesting Requires use of multi-genic approaches and precisely controlled gene expression

Benson Hill Biosystems – Company Summary • The Photosynthesis Company TM • Focus: Increased Crop Yield through Improved Photosynthesis • Discovery: Integrated, systems-based approach to engineering plant primary metabolism • One platform – multiple product opportunities: corn, sugarcane, soybean, rice, wheat, cotton, oil palm, canola, Eucalyptus, … • GM- and non-GM product concepts • World-class plant growth and genomics facility licensed and in use • Pre-Series A company; more partnership revenues than venture capital • Partnerships with global leaders

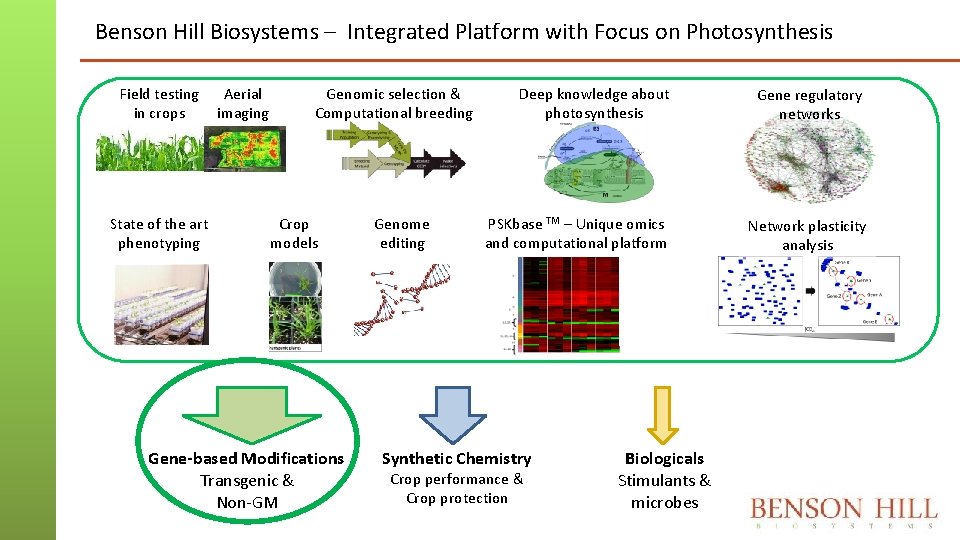
Benson Hill Biosystems – Integrated Platform with Focus on Photosynthesis Field testing in crops State of the art phenotyping Aerial imaging Genomic selection & Computational breeding Crop models Gene-based Modifications Transgenic & Non-GM Genome editing Deep knowledge about photosynthesis Gene regulatory networks PSKbase TM – Unique omics and computational platform Network plasticity analysis Synthetic Chemistry Crop performance & Crop protection Biologicals Stimulants & microbes

Partnership with Donald Danforth Plant Science Center • IP/license access to labs of Tom Brutnell and Todd Mockler – 35+ FTEs working on photosynthesis, computational and systems biology, transcriptional regulatory networks, novel targets and promoters, etc. • Enablement of Cap. Ex Lite with access to: Tissue Culture and Transformation Facility (2, 000 ft 2) Potting area with soils handling room (1, 900 ft 2) 33 Conviron chambers and 18 Conviron rooms (3, 000 ft 2) 36 Greenhouses (44, 000 ft 2) Bioinformatics Core: 800+ processors, 3 TB memory, and a single, high-performance 204 TB storage area network – Other Cores: Proteomics, Mass Spectrometry, Integrated Microscopy, X-Ray Crystallography – High-throughput robotics assay platforms – High-throughput plant phenotyping system – – –

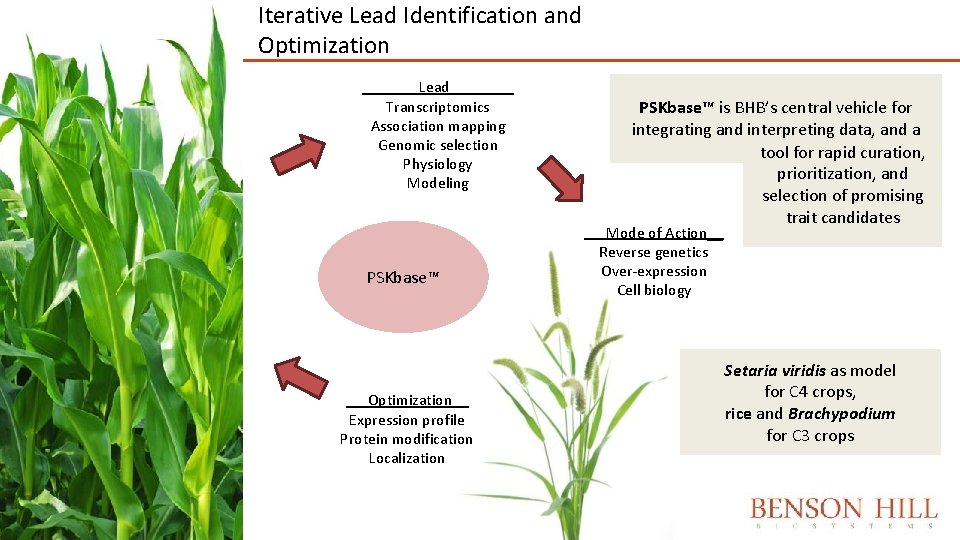
Iterative Lead Identification and Optimization ___ Lead____ Transcriptomics Association mapping Genomic selection Physiology Modeling PSKbase™ Optimization__ Expression profile Protein modification Localization PSKbase™ is BHB’s central vehicle for integrating and interpreting data, and a tool for rapid curation, prioritization, and selection of promising trait candidates Mode of Action__ Reverse genetics Over-expression Cell biology Setaria viridis as model for C 4 crops, rice and Brachypodium for C 3 crops

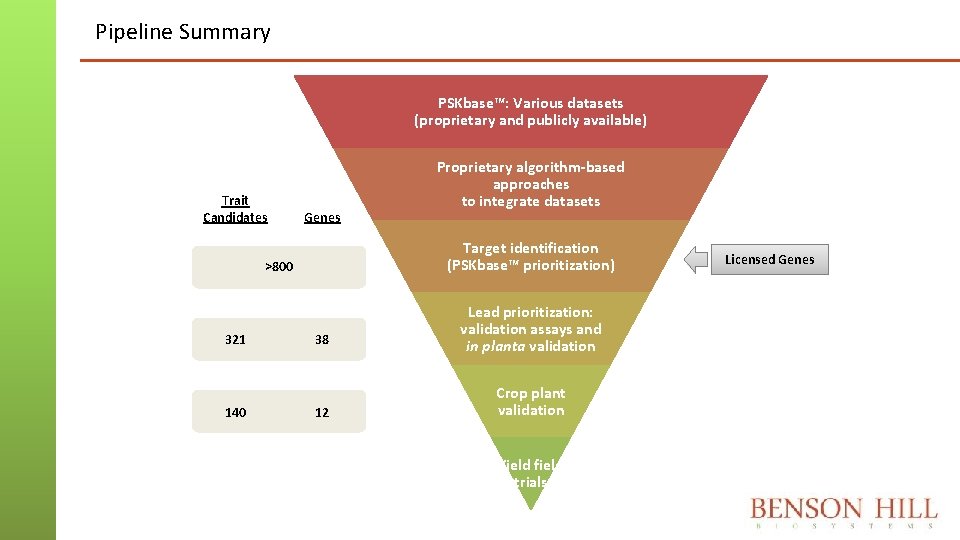
Pipeline Summary PSKbase™: Various datasets (proprietary and publicly available) Trait Candidates Genes Target identification (PSKbase™ prioritization) >800 321 140 Proprietary algorithm-based approaches to integrate datasets 38 12 Lead prioritization: validation assays and in planta validation Crop plant validation Yield field trials Licensed Genes

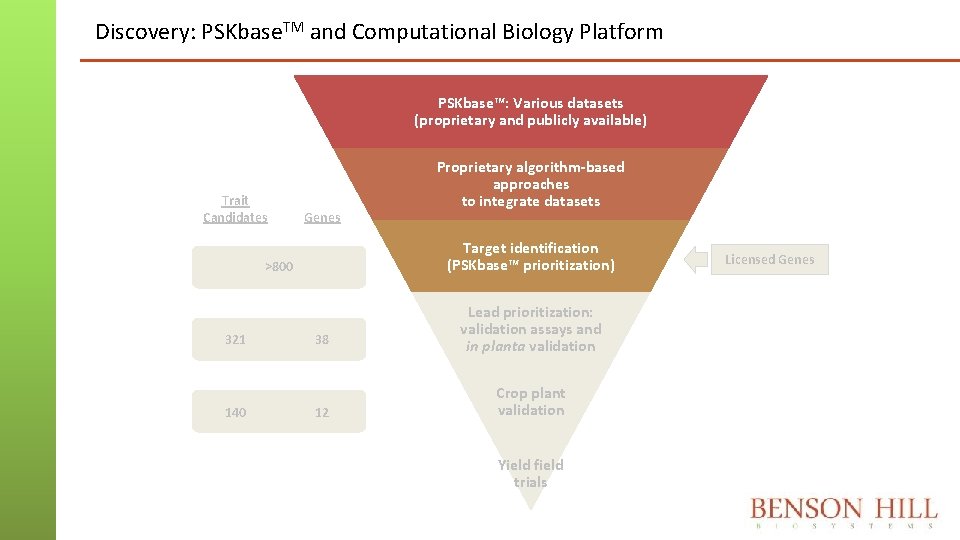
Discovery: PSKbase. TM and Computational Biology Platform PSKbase™: Various datasets (proprietary and publicly available) Trait Candidates Genes Target identification (PSKbase™ prioritization) >800 321 140 Proprietary algorithm-based approaches to integrate datasets 38 12 Lead prioritization: validation assays and in planta validation Crop plant validation Yield field trials Licensed Genes

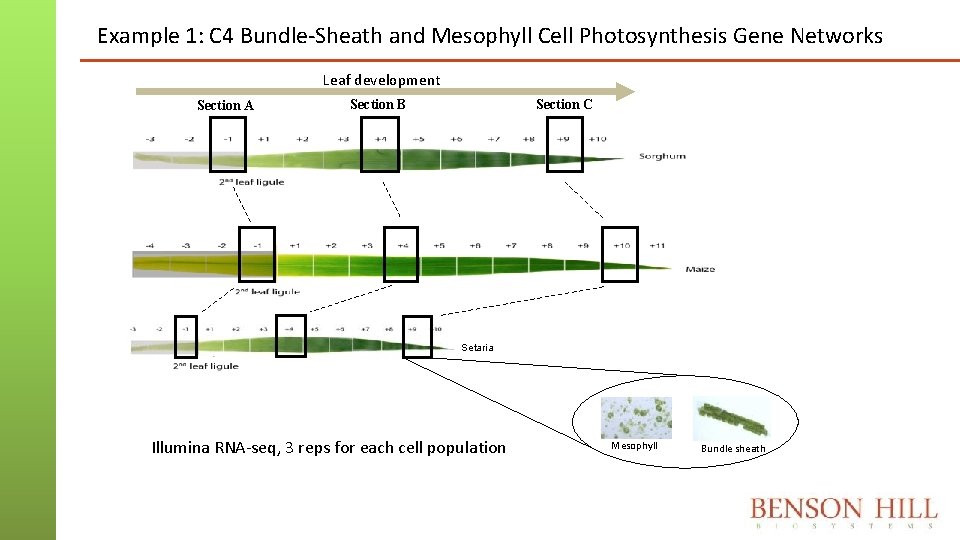
Example 1: C 4 Bundle-Sheath and Mesophyll Cell Photosynthesis Gene Networks Leaf development Section A Section C Section B Setaria Illumina RNA-seq, 3 reps for each cell population Mesophyll Bundle sheath
![Example 2 Wheat Leaf CO 2 responsive Gene Expression Atlas Low CO 2 Ambient Example 2: Wheat Leaf CO 2 -responsive Gene Expression Atlas Low [CO 2] Ambient](https://slidetodoc.com/presentation_image/5107c0415a18a3ad6316d0f65bad5255/image-16.jpg)
Example 2: Wheat Leaf CO 2 -responsive Gene Expression Atlas Low [CO 2] Ambient [CO 2] High [CO 2] – responsive functional gene candidates baseline developmental and metabolic gene expression

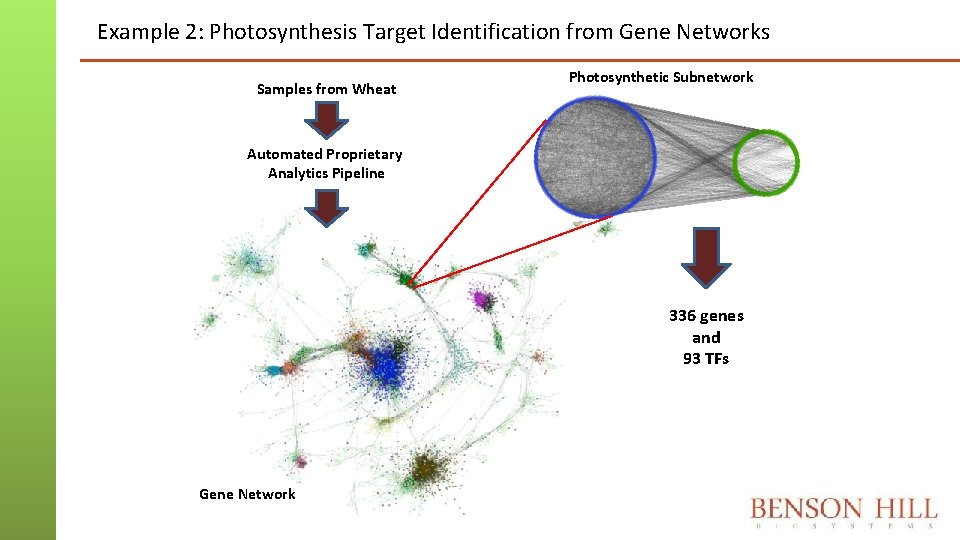
Example 2: Photosynthesis Target Identification from Gene Networks Samples from Wheat Photosynthetic Subnetwork Automated Proprietary Analytics Pipeline 336 genes and 93 TFs Gene Network

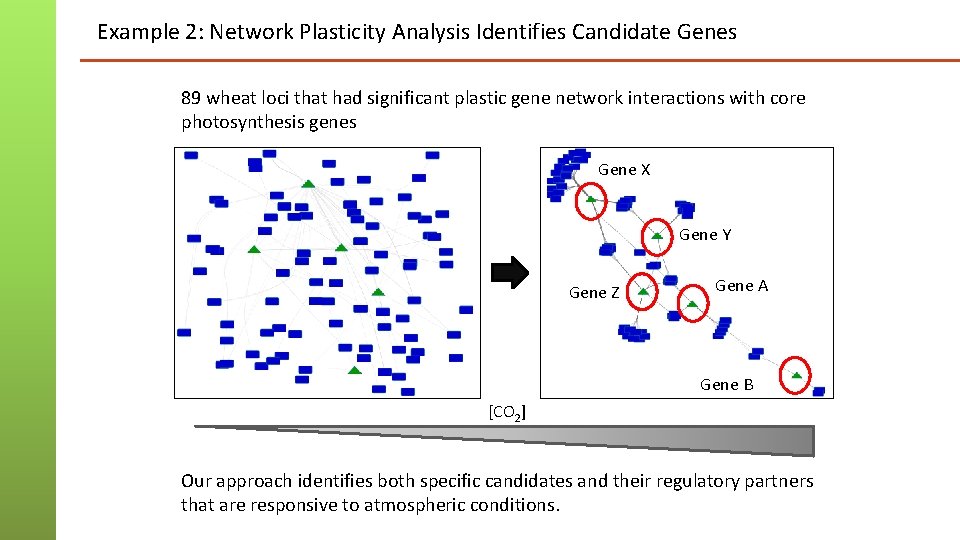
Example 2: Network Plasticity Analysis Identifies Candidate Genes 89 wheat loci that had significant plastic gene network interactions with core photosynthesis genes Gene X Gene Y Gene Z Gene A Gene B [CO 2] Our approach identifies both specific candidates and their regulatory partners that are responsive to atmospheric conditions.

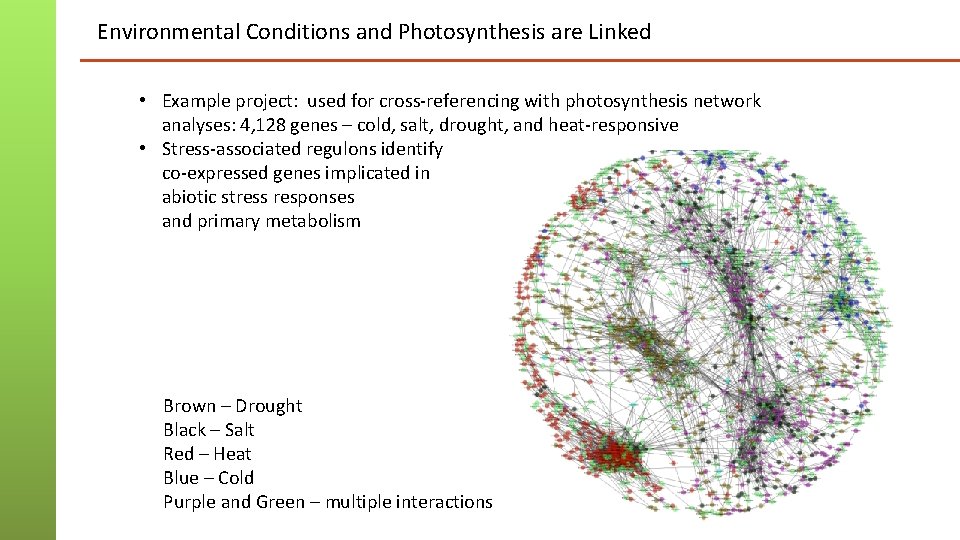
Environmental Conditions and Photosynthesis are Linked • Example project: used for cross-referencing with photosynthesis network analyses: 4, 128 genes – cold, salt, drought, and heat-responsive • Stress-associated regulons identify co-expressed genes implicated in abiotic stress responses and primary metabolism Brown – Drought Black – Salt Red – Heat Blue – Cold Purple and Green – multiple interactions

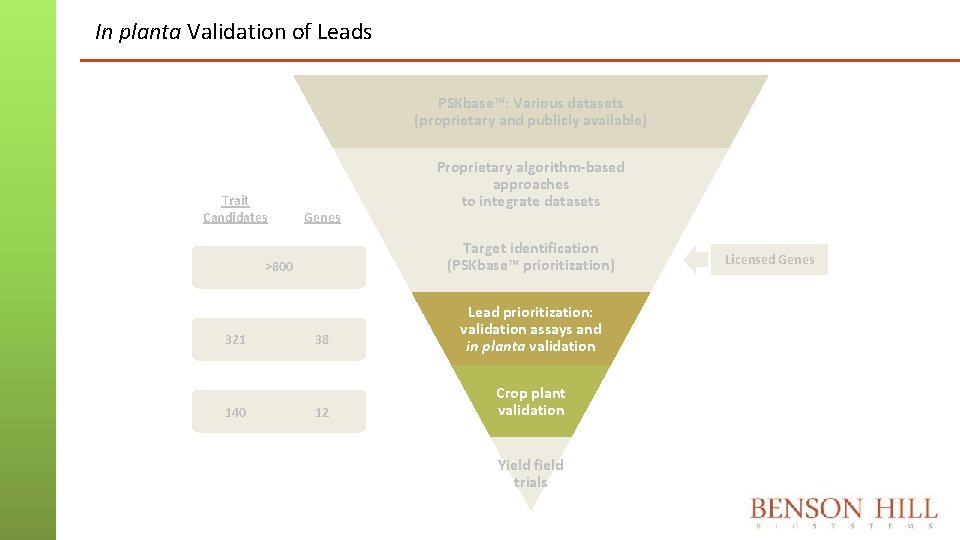
In planta Validation of Leads PSKbase™: Various datasets (proprietary and publicly available) Trait Candidates Genes Target identification (PSKbase™ prioritization) >800 321 140 Proprietary algorithm-based approaches to integrate datasets 38 12 Lead prioritization: validation assays and in planta validation Crop plant validation Yield field trials Licensed Genes

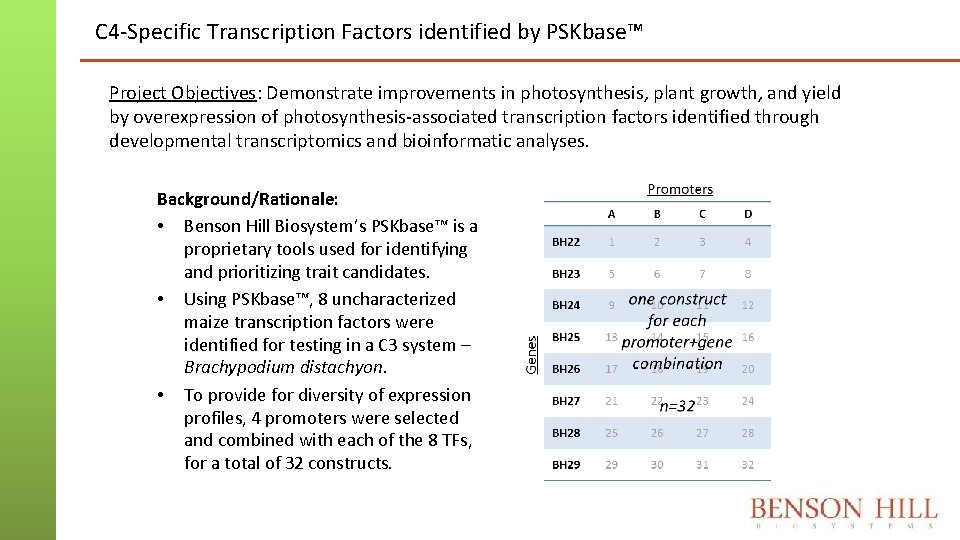
C 4 -Specific Transcription Factors identified by PSKbase™ Project Objectives: Demonstrate improvements in photosynthesis, plant growth, and yield by overexpression of photosynthesis-associated transcription factors identified through developmental transcriptomics and bioinformatic analyses. Background/Rationale: • Benson Hill Biosystem’s PSKbase™ is a proprietary tools used for identifying and prioritizing trait candidates. • Using PSKbase™, 8 uncharacterized maize transcription factors were identified for testing in a C 3 system – Brachypodium distachyon. • To provide for diversity of expression profiles, 4 promoters were selected and combined with each of the 8 TFs, for a total of 32 constructs.

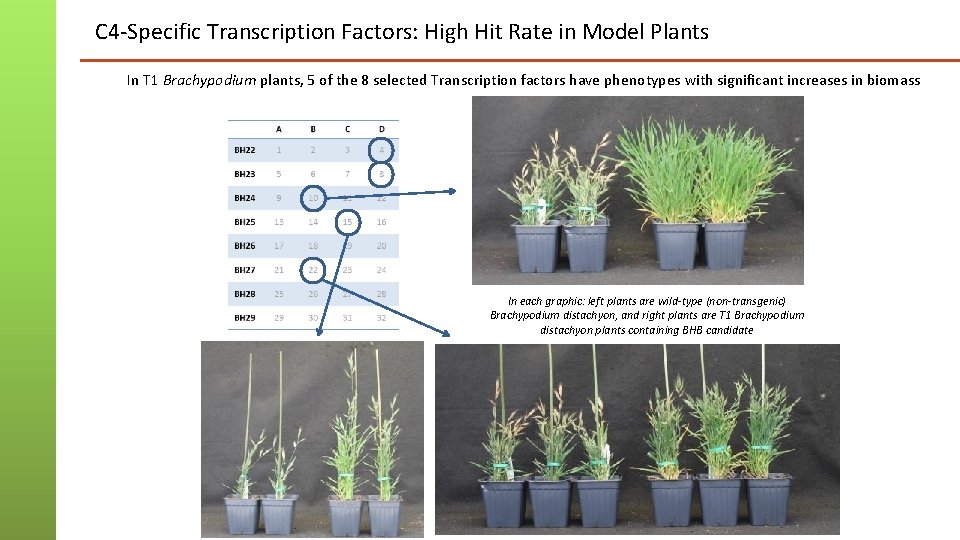
C 4 -Specific Transcription Factors: High Hit Rate in Model Plants In T 1 Brachypodium plants, 5 of the 8 selected Transcription factors have phenotypes with significant increases in biomass In each graphic: left plants are wild-type (non-transgenic) Brachypodium distachyon, and right plants are T 1 Brachypodium distachyon plants containing BHB candidate

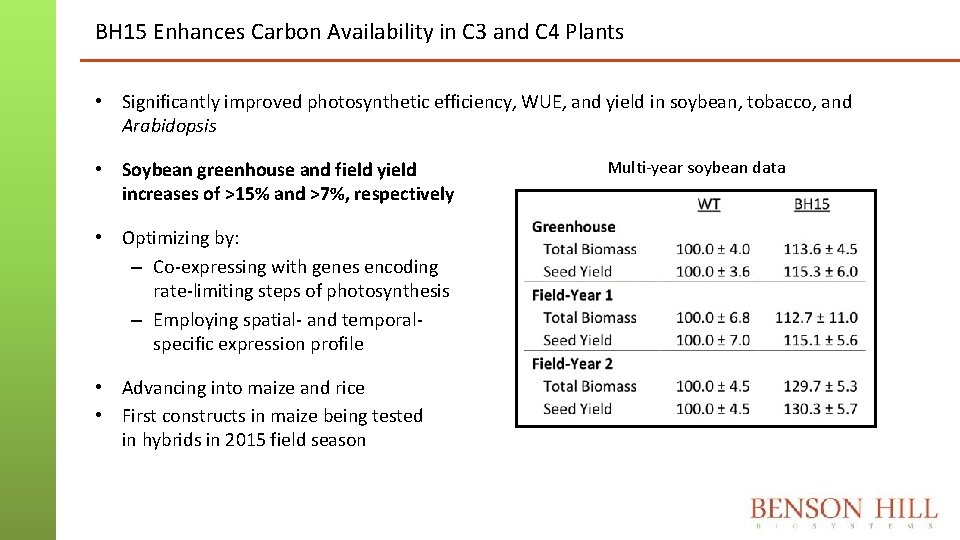
BH 15 Enhances Carbon Availability in C 3 and C 4 Plants • Significantly improved photosynthetic efficiency, WUE, and yield in soybean, tobacco, and Arabidopsis • Soybean greenhouse and field yield increases of >15% and >7%, respectively • Optimizing by: – Co-expressing with genes encoding rate-limiting steps of photosynthesis – Employing spatial- and temporalspecific expression profile • Advancing into maize and rice • First constructs in maize being tested in hybrids in 2015 field season Multi-year soybean data

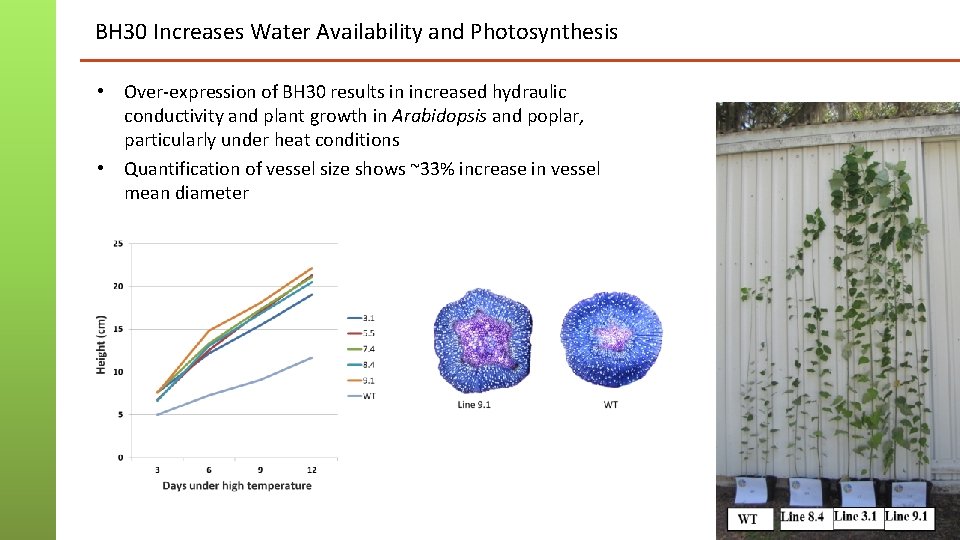
BH 30 Increases Water Availability and Photosynthesis • Over-expression of BH 30 results in increased hydraulic conductivity and plant growth in Arabidopsis and poplar, particularly under heat conditions • Quantification of vessel size shows ~33% increase in vessel mean diameter

BH 33 is a Well Characterized Sink Strength Lead • Naturally-occurring, well characterized enzyme in maize grain, mutated for improved thermal stability and enzyme kinetics • Previous mutated versions have shown promise in field trials of multiple crops • Iterative mutation has resulted in BH 33, which has improved characteristics relative to enzymes previously expressed in plants • Maize field testing in 2014 showed up to 24% increase in ear weight relative to control (null segregants with otherwise identical genetic background) • Inbred data in greenhouse also show positive results • Hybrid seed production completed and field trials in 2015 season ongoing • Coupling with numerous other source-focused trait candidates • Testing initiated in multiple other crops

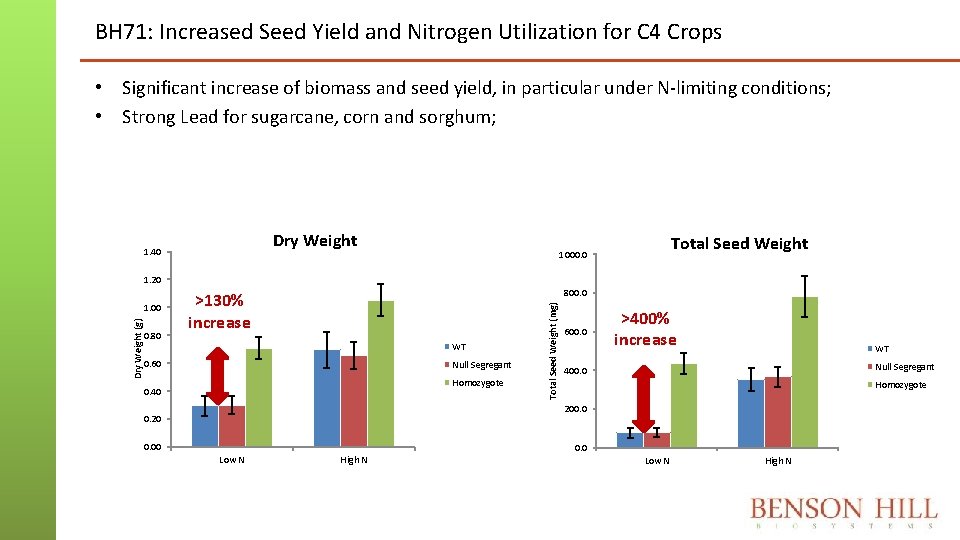
BH 71: Increased Seed Yield and Nitrogen Utilization for C 4 Crops • Significant increase of biomass and seed yield, in particular under N-limiting conditions; • Strong Lead for sugarcane, corn and sorghum; Dry Weight 1. 40 Total Seed Weight 1000. 0 1. 20 0. 80 800. 0 >130% increase WT 0. 60 Null Segregant Homozygote 0. 40 Total Seed Weight (mg) Dry Weight (g) 1. 00 600. 0 >400% increase WT Null Segregant 400. 0 Homozygote 200. 0 0. 20 0. 00 Low N High N 0. 0 Low N High N

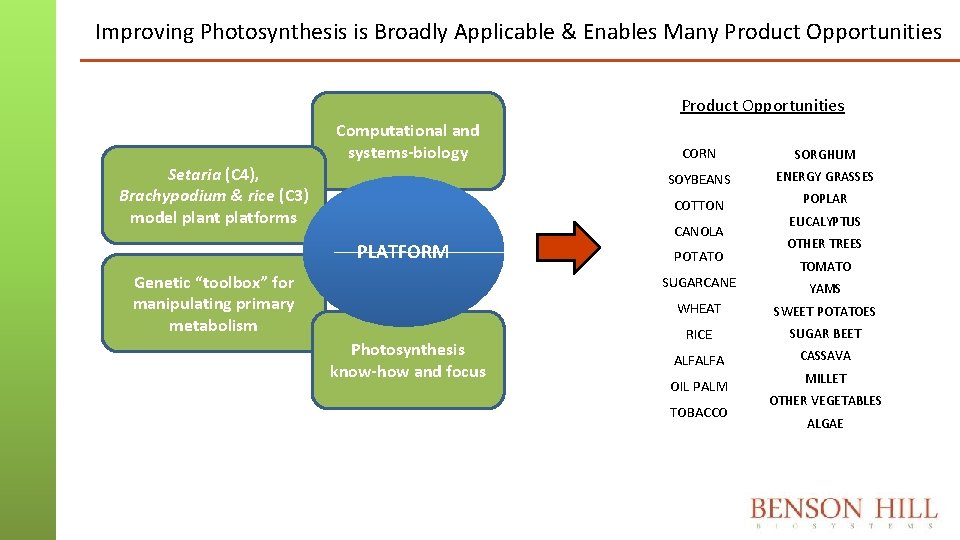
Improving Photosynthesis is Broadly Applicable & Enables Many Product Opportunities Setaria (C 4), Brachypodium & rice (C 3) model plant platforms Computational and systems-biology PLATFORM Genetic “toolbox” for manipulating primary metabolism CORN SORGHUM SOYBEANS ENERGY GRASSES COTTON POPLAR CANOLA POTATO SUGARCANE Photosynthesis know-how and focus EUCALYPTUS OTHER TREES TOMATO YAMS WHEAT SWEET POTATOES RICE SUGAR BEET ALFALFA CASSAVA OIL PALM TOBACCO MILLET OTHER VEGETABLES ALGAE

Path to Commercialization – “Go to Partner” • Seed market is consolidated, with high barriers to entry – Elite germplasm, i. e. plant genetics REQUIRES PARTNERSHIPS • Biotech trait discovery and development is entry point for participating in the most valuable and high-growth segment • To monetize traits, seed are used as value capture mechanism – Premium pricing for seed containing biotech trait(s) – Value sharing via royalties, which can be pre-calculated (flat rate) or based on percentage of trait value retained by seed company • Benson Hill Biosystems is partnering on a crop-by-crop basis – Non-exclusive deal structures in corn and soybean – Exclusive and alternative deal structures in other crops

Thank You

Non-GM Product Concept: Genome Editing Technology Prerequisite: most BHB lead genes are derived from crops Validate targets identified through PSKbase™ Platform agnostic: CRISPR/Cas, TALEN, Meganucleases Precise introduction of foreign genes Modification of target gene expression Replacement of genes by improved versions Uniform expression levels (only few events required) Modification of native genes through small insertions – Regulatory motifs to modify gene expression – Specific base pair changes to modify activity/specificity of native enzymes • Potential Non-GMO approach – Of particular relevance for wheat, rice, etc. • USDA confirms BHB’s approach as non-regulated, i. e. non-GM products • •