Incorporating uncertainty in distancematrix phylogenetics Wally Gilks Tom




- Slides: 14

Incorporating uncertainty in distance-matrix phylogenetics Wally Gilks Tom Nye Pietro Liò Leeds University Newcastle University Cambridge University Isaac Newton Institute December 17, 2007

Distance-based methods • Larger trees • Faster algorithms • Less model-dependent – Genome-scale evolutionary rearrangements

Agglomerative distance methods • NJ (Saitou and Nei, 1987) • Bio. NJ (Gascuel, 1997) • Weighbor (Bruno et al, 2000) • MVR (Gascuel, 2000) • Fast. ME (Desper and Gascuel, 2004)

Variance models • Independent distances – Ordinary Least Squares (OLS) – Weighted Least Squares (WLS) A – NJ, Weighbor, Fast. ME • Correlated distances – – shared evolutionary paths (Chakraborty, 1977) computed from shared sequences: Bio. NJ induced by estimation process (we show) Generalised Least Squares (GLS) – Hasegawa (1985), Bulmer (1991), MVR A B C

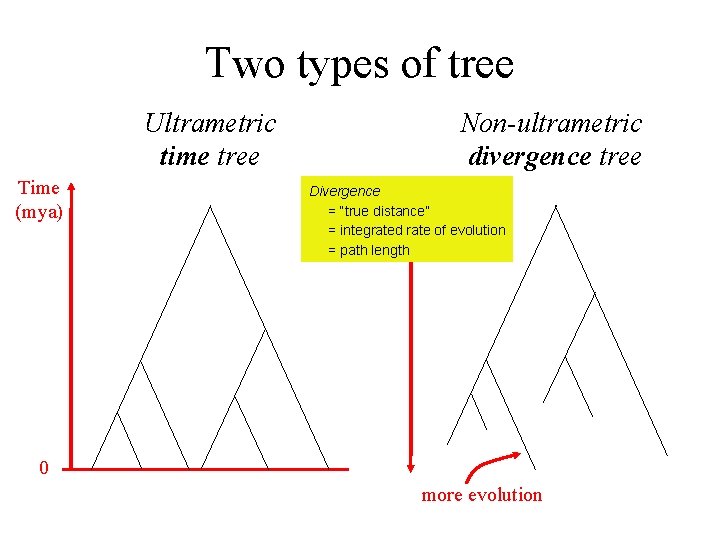
Two types of tree Ultrametric time tree Time (mya) Non-ultrametric divergence tree Divergence = “true distance” = integrated rate of evolution = path length 0 more evolution

Which tree type to assume? • • Ultrametric tree makes stronger assumptions Different methods for estimating each type But both types are in principle correct! Our method coherently integrates both types – Produces rooted tree, no need for outgroup

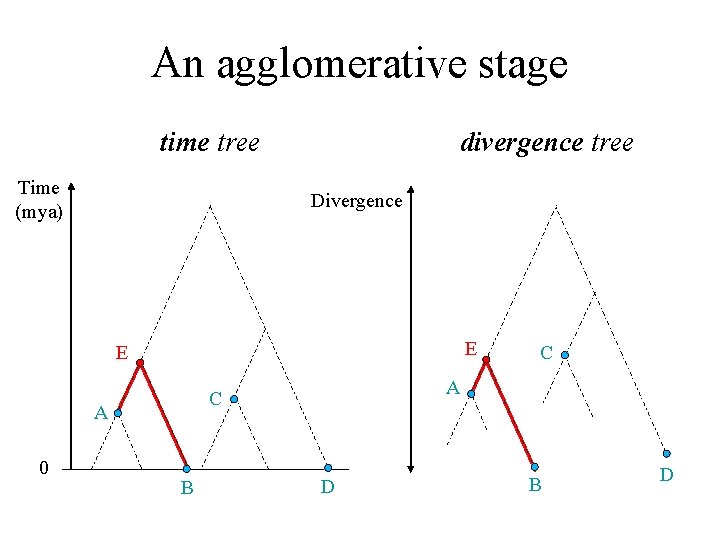
An agglomerative stage time tree Time (mya) divergence tree Divergence E E 0 A C A B C D B D

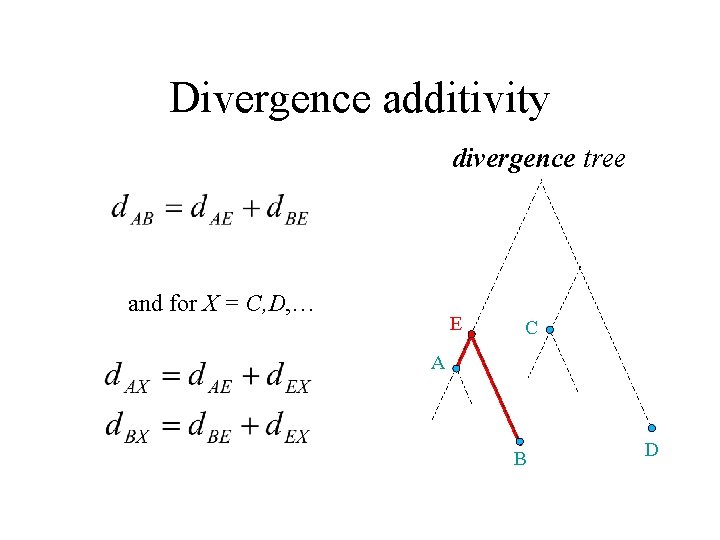
Divergence additivity divergence tree and for X = C, D, … E C A B D

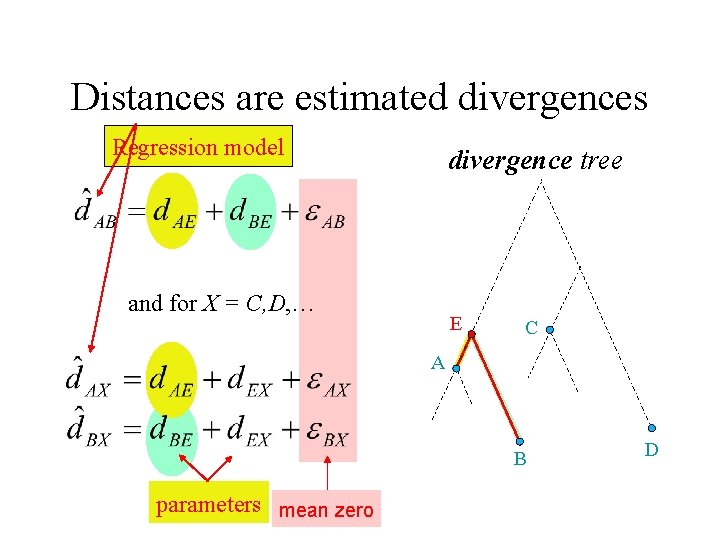
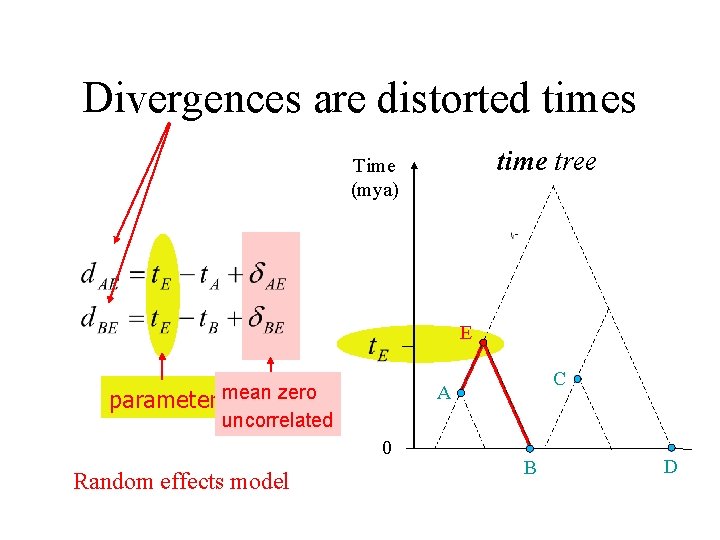
Distances are estimated divergences Regression model divergence tree and for X = C, D, … E C A B parameters mean zero D

Divergences are distorted times time tree Time (mya) E parameter mean zero uncorrelated 0 Random effects model C A B D

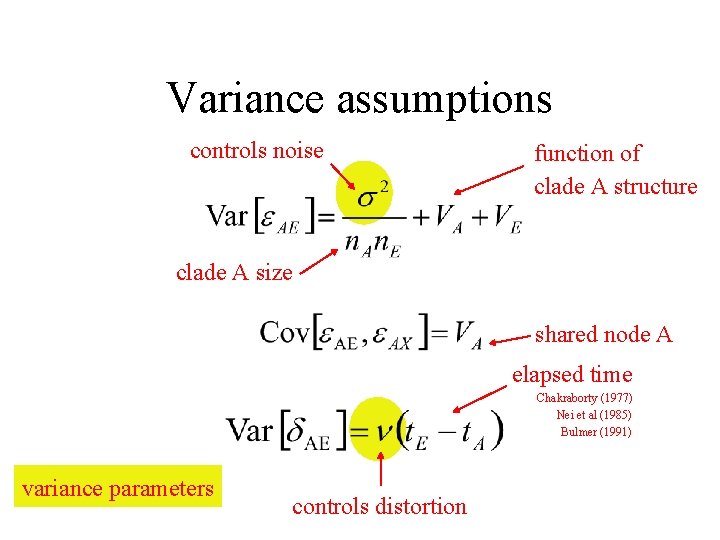
Variance assumptions controls noise function of clade A structure clade A size shared node A elapsed time Chakraborty (1977) Nei et al (1985) Bulmer (1991) variance parameters controls distortion

Estimation • Time tree and divergence tree are estimated simultaneously – by GLS (Hasegawa, 1985; Bulmer, 1991) • Choose most recent agglomeration always • Estimated divergences become the distances for the next stage – Variance formula accommodates estimation-induced correlations

Notes • Can estimate variance parameters s 2 and n • Computationally efficient algorithm – same time-complexity as Bio. NJ – we call it Stat. Tree

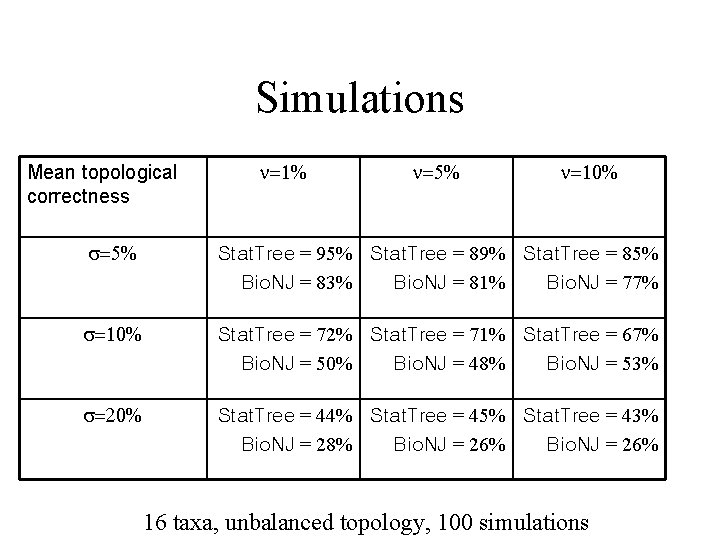
Simulations Mean topological correctness n=1% n=5% n=10% s=5% Stat. Tree = 95% Stat. Tree = 89% Stat. Tree = 85% Bio. NJ = 83% Bio. NJ = 81% Bio. NJ = 77% s=10% Stat. Tree = 72% Stat. Tree = 71% Stat. Tree = 67% Bio. NJ = 50% Bio. NJ = 48% Bio. NJ = 53% s=20% Stat. Tree = 44% Stat. Tree = 45% Stat. Tree = 43% Bio. NJ = 28% Bio. NJ = 26% 16 taxa, unbalanced topology, 100 simulations
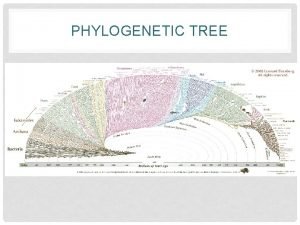
 Mega phylogenetics
Mega phylogenetics Biology taxonomy tree
Biology taxonomy tree Rooted vs unrooted phylogenetic tree
Rooted vs unrooted phylogenetic tree Incorporating pronunciation
Incorporating pronunciation Incorporating the change
Incorporating the change Incorporating in ohio
Incorporating in ohio Picking up and incorporating dna from dead bacterial cells
Picking up and incorporating dna from dead bacterial cells Capital budgeting chapter
Capital budgeting chapter Nested quotation example
Nested quotation example Walraven van hall monumento significado
Walraven van hall monumento significado Wally gibson
Wally gibson Sir walter wally
Sir walter wally Wally olins podręcznik brandingu
Wally olins podręcznik brandingu Dotation
Dotation Rph pathology
Rph pathology