Imprinting Expression of only one allele of a




- Slides: 32

Imprinting Expression of only one allele of a locus Only ~100 genes in mammals are imprinted Parthenogenesis is not possible in mammals due to incorrect expression of imprinted genes Most imprinted genes are involved in growth control or postnatal behavior Imprinted genes involves allele specific methylation and are resistant to genome-wide demethylation in germ cell development Some imprinted gene clusters are regulated by methylation-regulated insulators Some clusters of imprinted genes contain long nc. RNAs that control allele-specific expression

Kinship Theory of Imprinting Conflict exists between the interests of the paternal and maternal genes For optimal fitness of the father, paternal genes maximize acquisition of maternal resources to ensure larger sized offspring Maternal genes are sparing in the demands of maternal resources, so that the mother has a better chance to bear further offspring Paternally-expressed genes generally stimulate growth Maternally-expressed genes generally repress growth

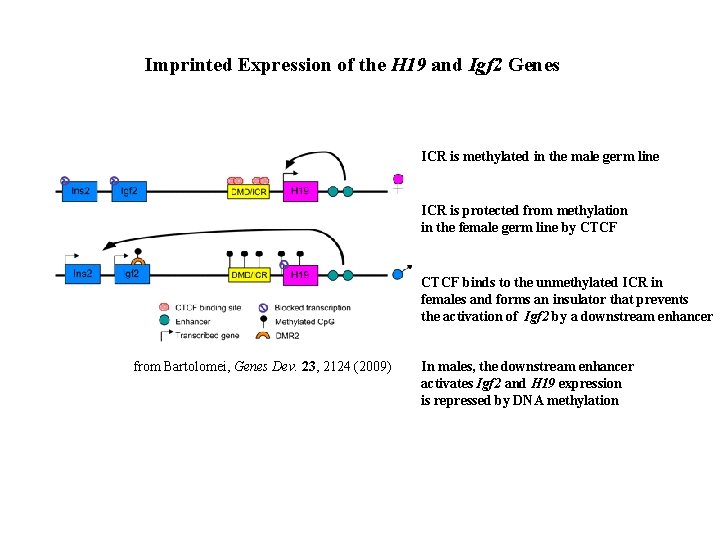
Imprinted Expression of the H 19 and Igf 2 Genes ICR is methylated in the male germ line ICR is protected from methylation in the female germ line by CTCF binds to the unmethylated ICR in females and forms an insulator that prevents the activation of Igf 2 by a downstream enhancer from Bartolomei, Genes Dev. 23, 2124 (2009) In males, the downstream enhancer activates Igf 2 and H 19 expression is repressed by DNA methylation

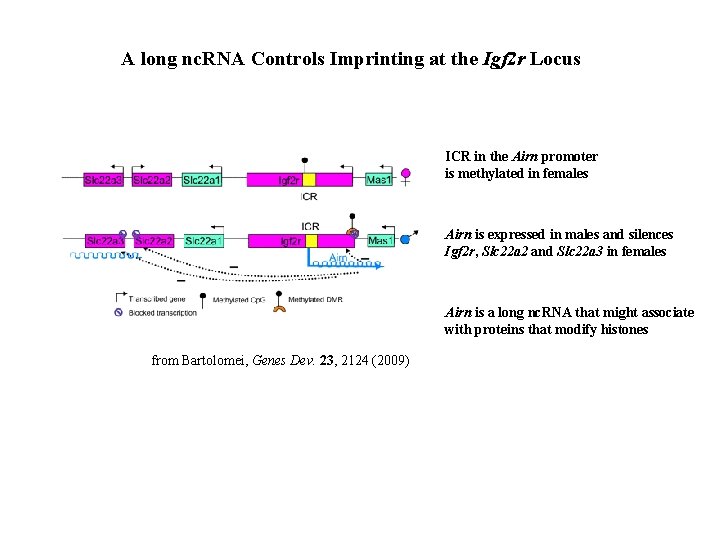
A long nc. RNA Controls Imprinting at the Igf 2 r Locus ICR in the Airn promoter is methylated in females Airn is expressed in males and silences Igf 2 r, Slc 22 a 2 and Slc 22 a 3 in females Airn is a long nc. RNA that might associate with proteins that modify histones from Bartolomei, Genes Dev. 23, 2124 (2009)

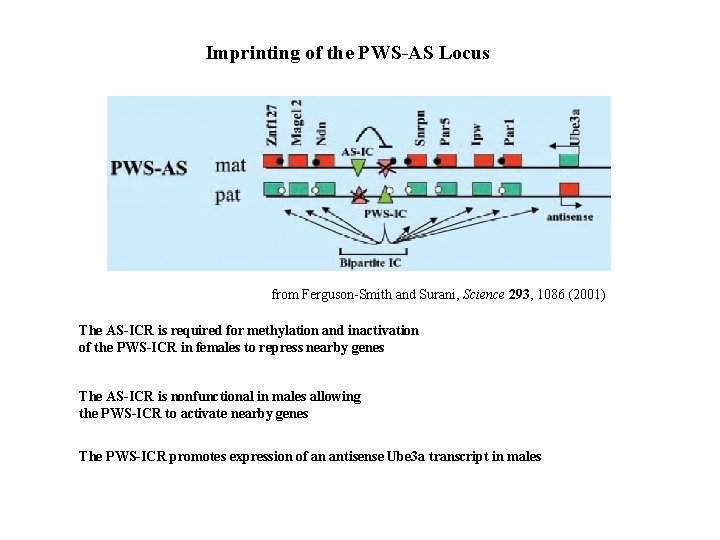
Imprinting of the PWS-AS Locus from Ferguson-Smith and Surani, Science 293, 1086 (2001) The AS-ICR is required for methylation and inactivation of the PWS-ICR in females to repress nearby genes The AS-ICR is nonfunctional in males allowing the PWS-ICR to activate nearby genes The PWS-ICR promotes expression of an antisense Ube 3 a transcript in males

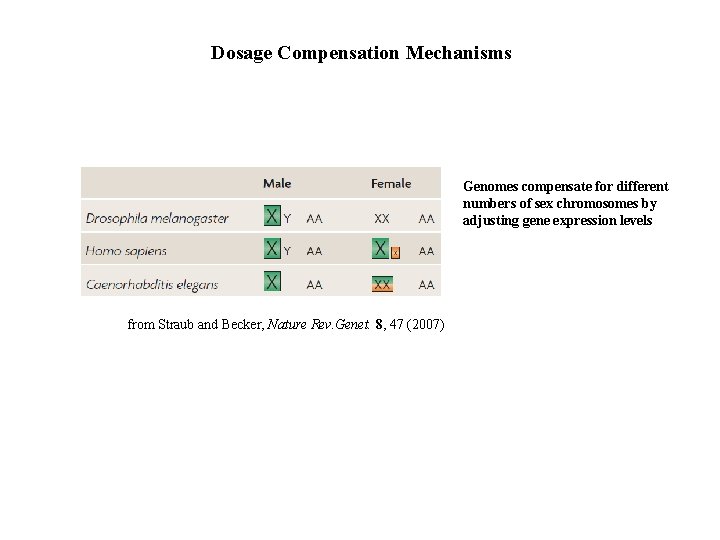
Dosage Compensation Mechanisms Genomes compensate for different numbers of sex chromosomes by adjusting gene expression levels from Straub and Becker, Nature Rev. Genet. 8, 47 (2007)

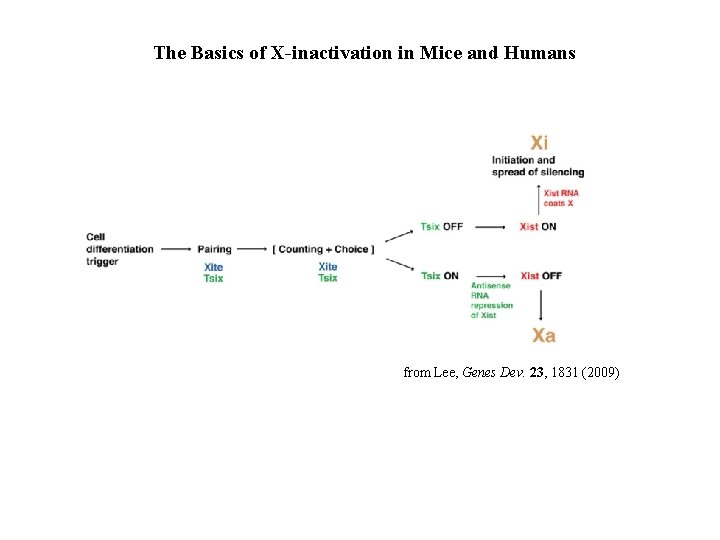
The Basics of X-inactivation in Mice and Humans from Lee, Genes Dev. 23, 1831 (2009)

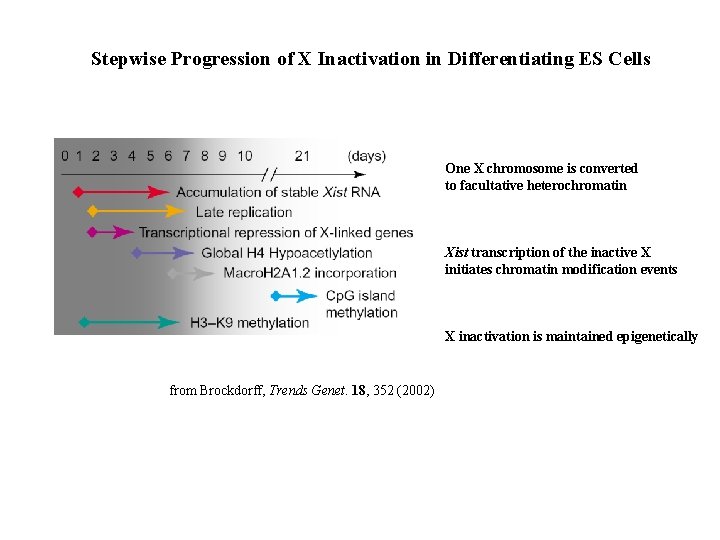
Stepwise Progression of X Inactivation in Differentiating ES Cells One X chromosome is converted to facultative heterochromatin Xist transcription of the inactive X initiates chromatin modification events X inactivation is maintained epigenetically from Brockdorff, Trends Genet. 18, 352 (2002)

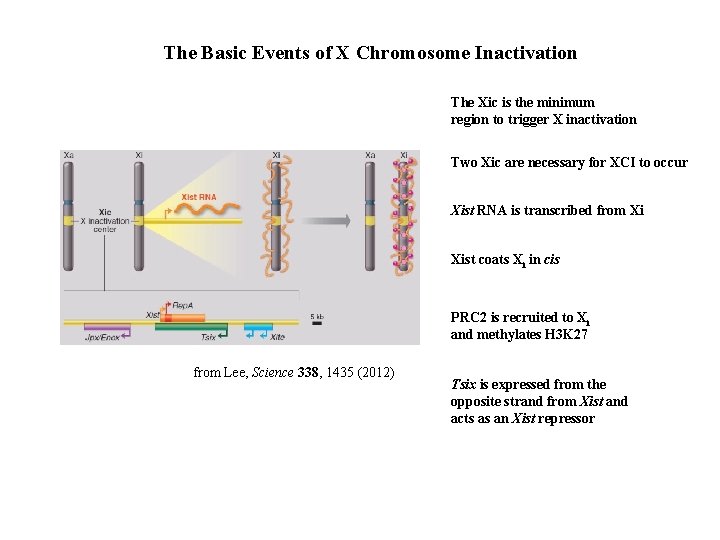
The Basic Events of X Chromosome Inactivation The Xic is the minimum region to trigger X inactivation Two Xic are necessary for XCI to occur Xist RNA is transcribed from Xi Xist coats Xi in cis PRC 2 is recruited to Xi and methylates H 3 K 27 from Lee, Science 338, 1435 (2012) Tsix is expressed from the opposite strand from Xist and acts as an Xist repressor

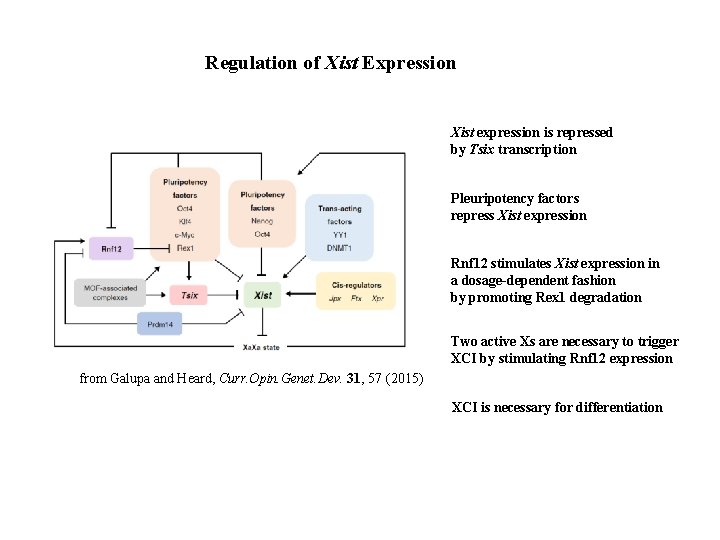
Regulation of Xist Expression Xist expression is repressed by Tsix transcription Pleuripotency factors repress Xist expression Rnf 12 stimulates Xist expression in a dosage-dependent fashion by promoting Rex 1 degradation Two active Xs are necessary to trigger XCI by stimulating Rnf 12 expression from Galupa and Heard, Curr. Opin. Genet. Dev. 31, 57 (2015) XCI is necessary for differentiation

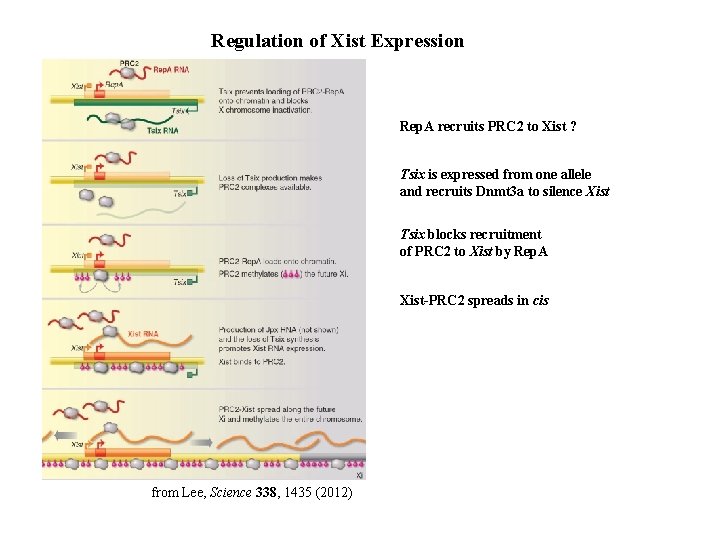
Regulation of Xist Expression Rep. A recruits PRC 2 to Xist ? Tsix is expressed from one allele and recruits Dnmt 3 a to silence Xist Tsix blocks recruitment of PRC 2 to Xist by Rep. A Xist-PRC 2 spreads in cis from Lee, Science 338, 1435 (2012)

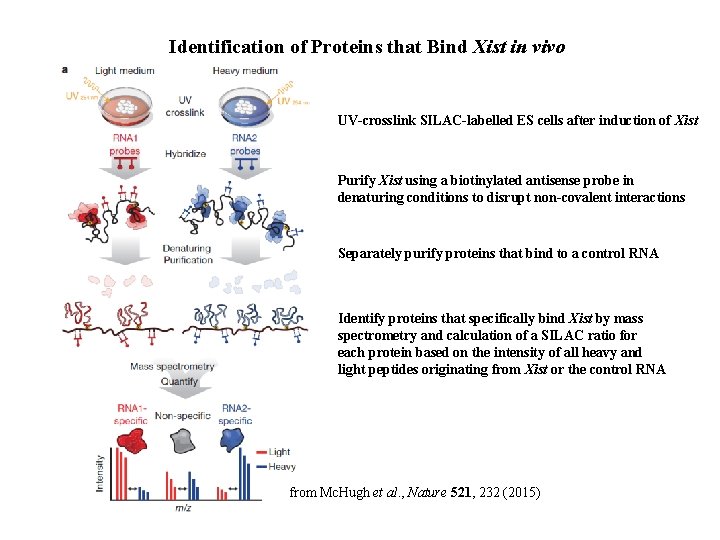
Identification of Proteins that Bind Xist in vivo UV-crosslink SILAC-labelled ES cells after induction of Xist Purify Xist using a biotinylated antisense probe in denaturing conditions to disrupt non-covalent interactions Separately purify proteins that bind to a control RNA Identify proteins that specifically bind Xist by mass spectrometry and calculation of a SILAC ratio for each protein based on the intensity of all heavy and light peptides originating from Xist or the control RNA from Mc. Hugh et al. , Nature 521, 232 (2015)

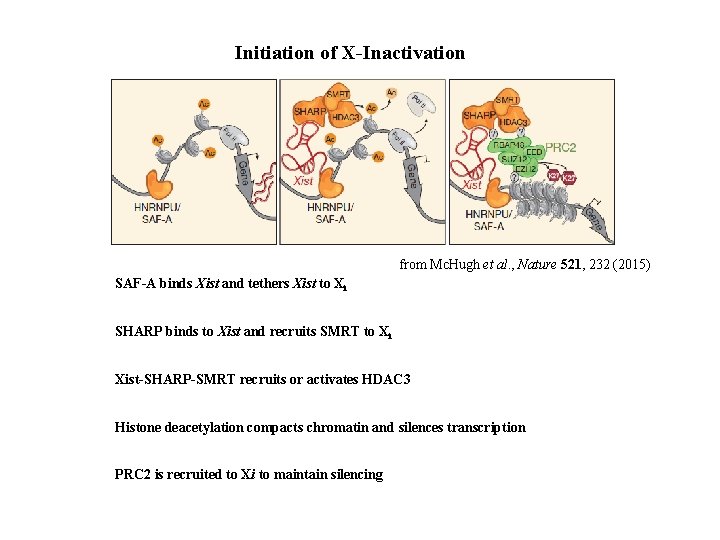
Initiation of X-Inactivation from Mc. Hugh et al. , Nature 521, 232 (2015) SAF-A binds Xist and tethers Xist to Xi SHARP binds to Xist and recruits SMRT to Xi Xist-SHARP-SMRT recruits or activates HDAC 3 Histone deacetylation compacts chromatin and silences transcription PRC 2 is recruited to Xi to maintain silencing

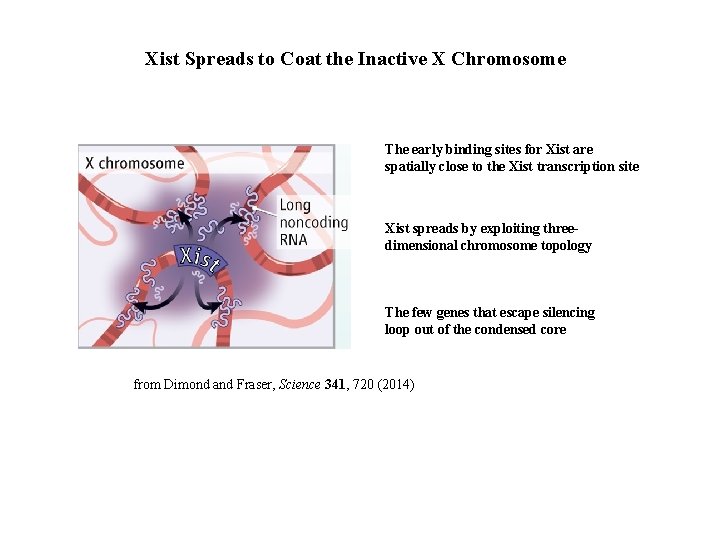
Xist Spreads to Coat the Inactive X Chromosome The early binding sites for Xist are spatially close to the Xist transcription site Xist spreads by exploiting threedimensional chromosome topology The few genes that escape silencing loop out of the condensed core from Dimond and Fraser, Science 341, 720 (2014)

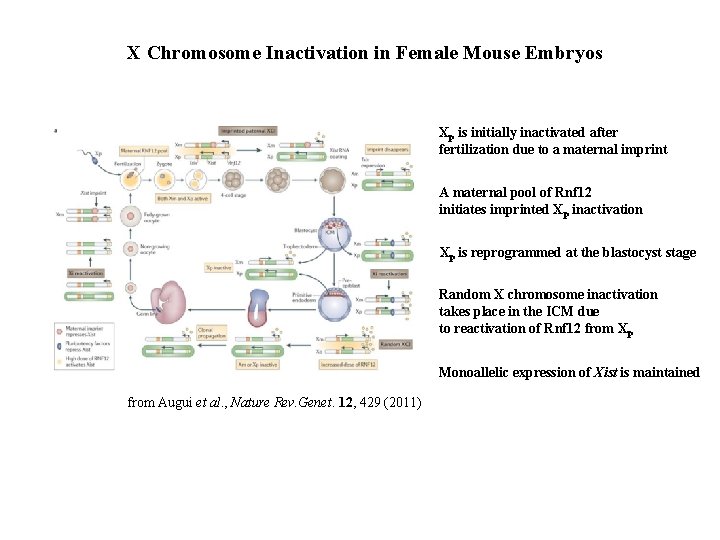
X Chromosome Inactivation in Female Mouse Embryos Xp is initially inactivated after fertilization due to a maternal imprint A maternal pool of Rnf 12 initiates imprinted Xp inactivation Xp is reprogrammed at the blastocyst stage Random X chromosome inactivation takes place in the ICM due to reactivation of Rnf 12 from Xp Monoallelic expression of Xist is maintained from Augui et al. , Nature Rev. Genet. 12, 429 (2011)

Calico Cats One of the genes controlling fur color is on the X chromosome B – orange b - black Female mammals are genetic mosaics Random X inactivation early in embryonic development leads to patchworks of skin cells expressing each allele

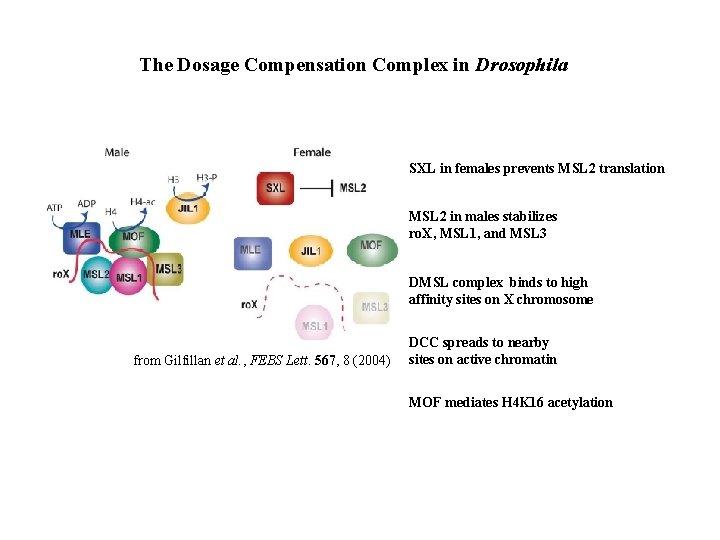
The Dosage Compensation Complex in Drosophila SXL in females prevents MSL 2 translation MSL 2 in males stabilizes ro. X, MSL 1, and MSL 3 DMSL complex binds to high affinity sites on X chromosome from Gilfillan et al. , FEBS Lett. 567, 8 (2004) DCC spreads to nearby sites on active chromatin MOF mediates H 4 K 16 acetylation

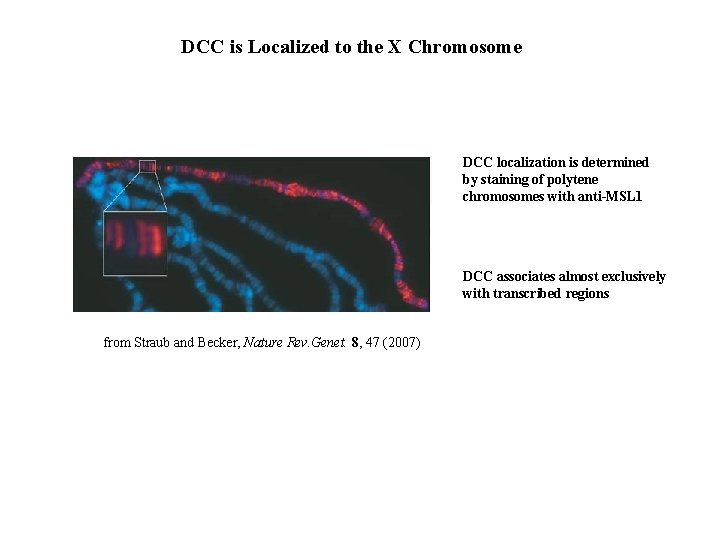
DCC is Localized to the X Chromosome DCC localization is determined by staining of polytene chromosomes with anti-MSL 1 DCC associates almost exclusively with transcribed regions from Straub and Becker, Nature Rev. Genet. 8, 47 (2007)

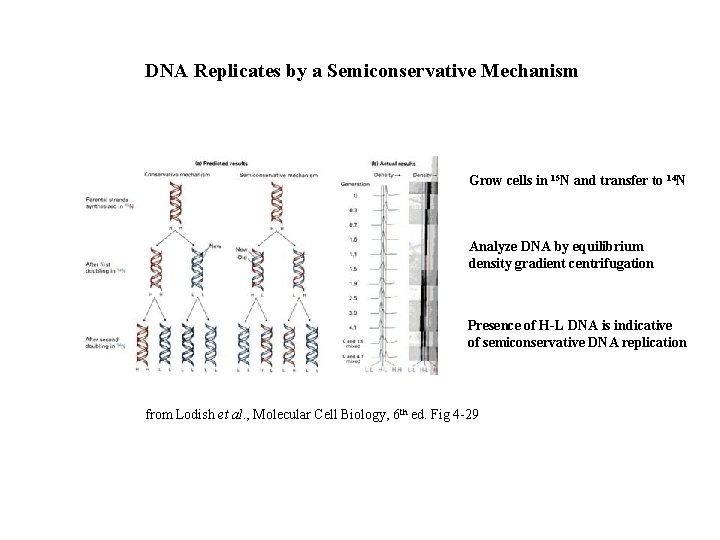
DNA Replicates by a Semiconservative Mechanism Grow cells in 15 N and transfer to 14 N Analyze DNA by equilibrium density gradient centrifugation Presence of H-L DNA is indicative of semiconservative DNA replication from Lodish et al. , Molecular Cell Biology, 6 th ed. Fig 4 -29

The 11 th Commandment

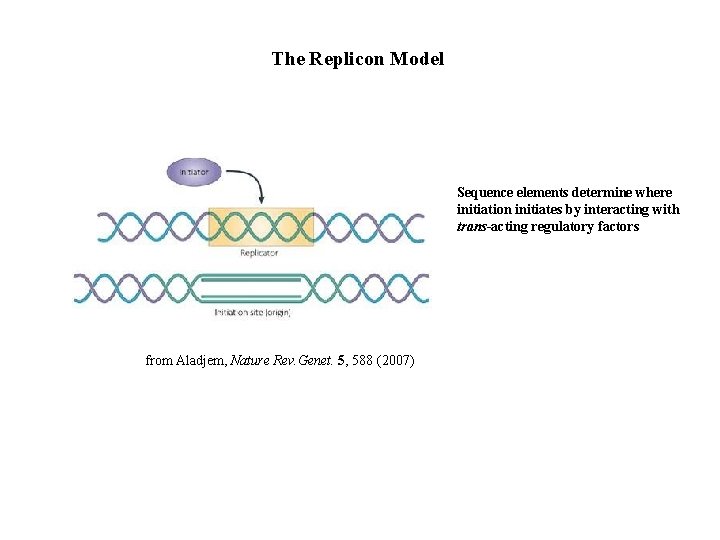
The Replicon Model Sequence elements determine where initiation initiates by interacting with trans-acting regulatory factors from Aladjem, Nature Rev. Genet. 5, 588 (2007)

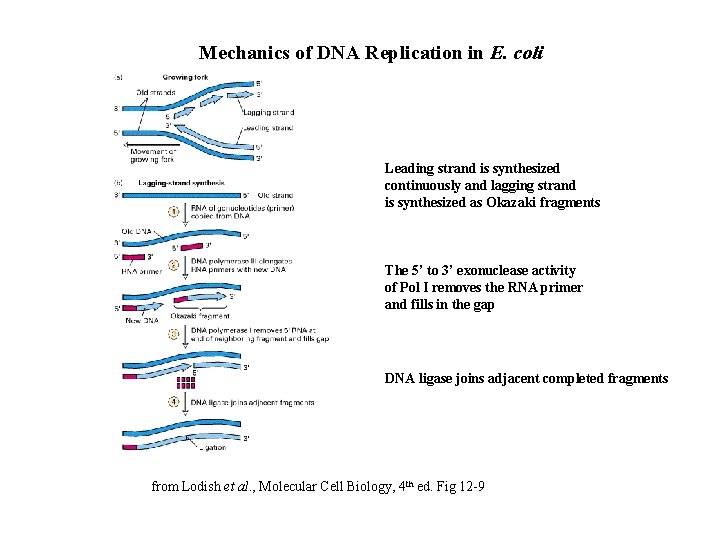
Mechanics of DNA Replication in E. coli Leading strand is synthesized continuously and lagging strand is synthesized as Okazaki fragments The 5’ to 3’ exonuclease activity of Pol I removes the RNA primer and fills in the gap DNA ligase joins adjacent completed fragments from Lodish et al. , Molecular Cell Biology, 4 th ed. Fig 12 -9

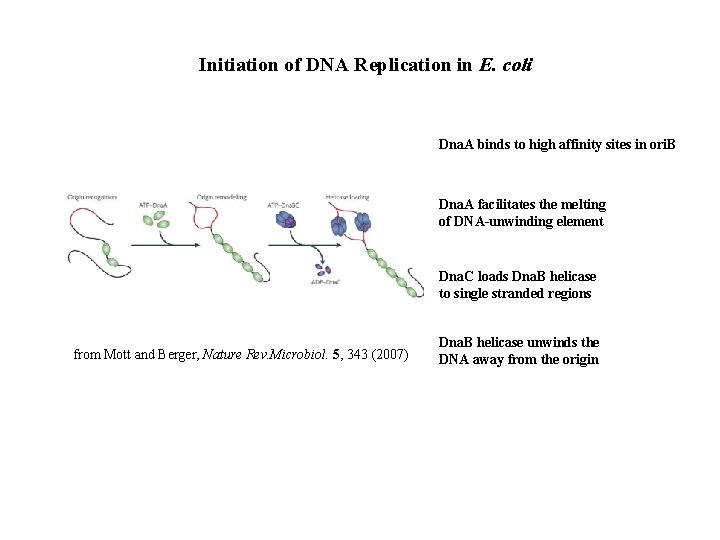
Initiation of DNA Replication in E. coli Dna. A binds to high affinity sites in ori. B Dna. A facilitates the melting of DNA-unwinding element Dna. C loads Dna. B helicase to single stranded regions from Mott and Berger, Nature Rev. Microbiol. 5, 343 (2007) Dna. B helicase unwinds the DNA away from the origin

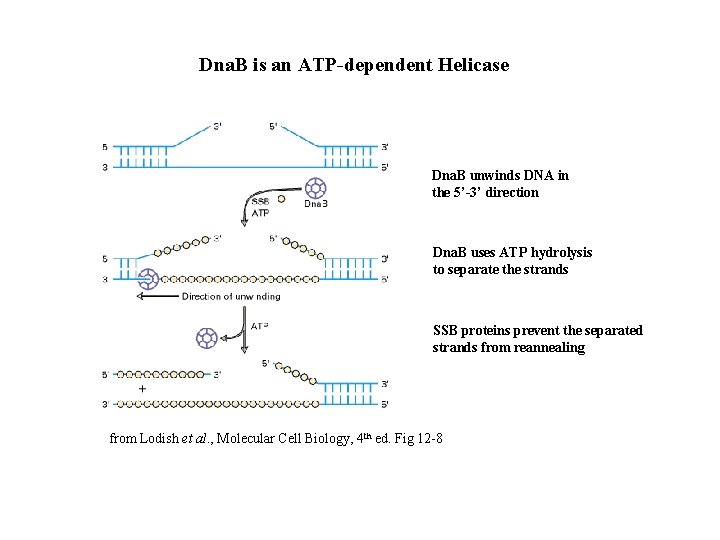
Dna. B is an ATP-dependent Helicase Dna. B unwinds DNA in the 5’-3’ direction Dna. B uses ATP hydrolysis to separate the strands SSB proteins prevent the separated strands from reannealing from Lodish et al. , Molecular Cell Biology, 4 th ed. Fig 12 -8

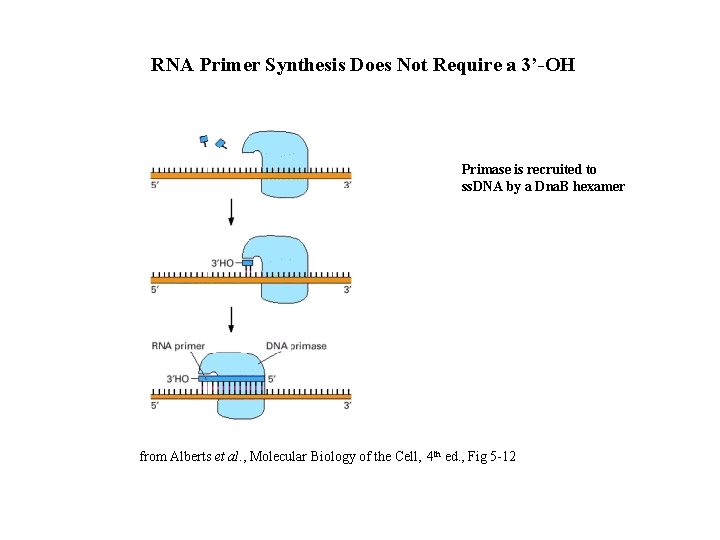
RNA Primer Synthesis Does Not Require a 3’-OH Primase is recruited to ss. DNA by a Dna. B hexamer from Alberts et al. , Molecular Biology of the Cell, 4 th ed. , Fig 5 -12

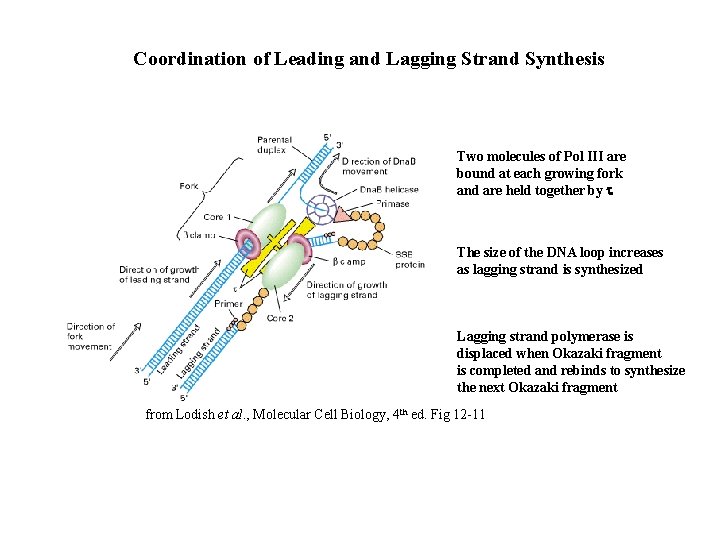
Coordination of Leading and Lagging Strand Synthesis Two molecules of Pol III are bound at each growing fork and are held together by t The size of the DNA loop increases as lagging strand is synthesized Lagging strand polymerase is displaced when Okazaki fragment is completed and rebinds to synthesize the next Okazaki fragment from Lodish et al. , Molecular Cell Biology, 4 th ed. Fig 12 -11

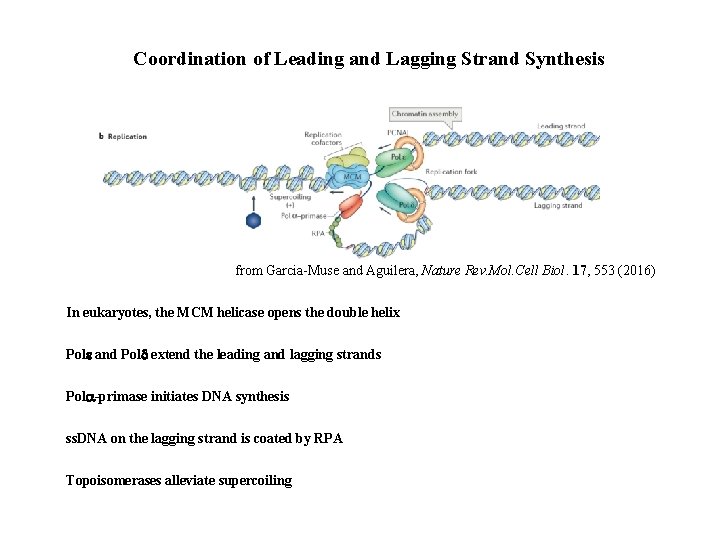
Coordination of Leading and Lagging Strand Synthesis from Garcia-Muse and Aguilera, Nature Rev. Mol. Cell Biol. 17, 553 (2016) In eukaryotes, the MCM helicase opens the double helix Pole and Pold extend the leading and lagging strands Pola-primase initiates DNA synthesis ss. DNA on the lagging strand is coated by RPA Topoisomerases alleviate supercoiling

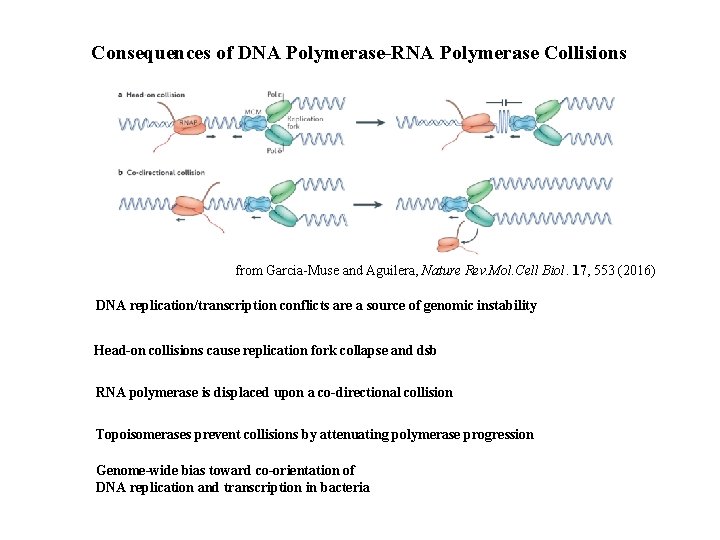
Consequences of DNA Polymerase-RNA Polymerase Collisions from Garcia-Muse and Aguilera, Nature Rev. Mol. Cell Biol. 17, 553 (2016) DNA replication/transcription conflicts are a source of genomic instability Head-on collisions cause replication fork collapse and dsb RNA polymerase is displaced upon a co-directional collision Topoisomerases prevent collisions by attenuating polymerase progression Genome-wide bias toward co-orientation of DNA replication and transcription in bacteria

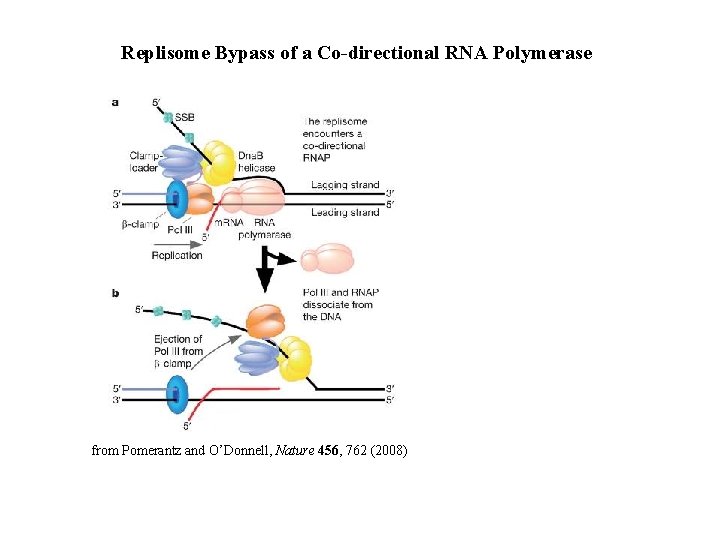
Replisome Bypass of a Co-directional RNA Polymerase from Pomerantz and O’Donnell, Nature 456, 762 (2008)

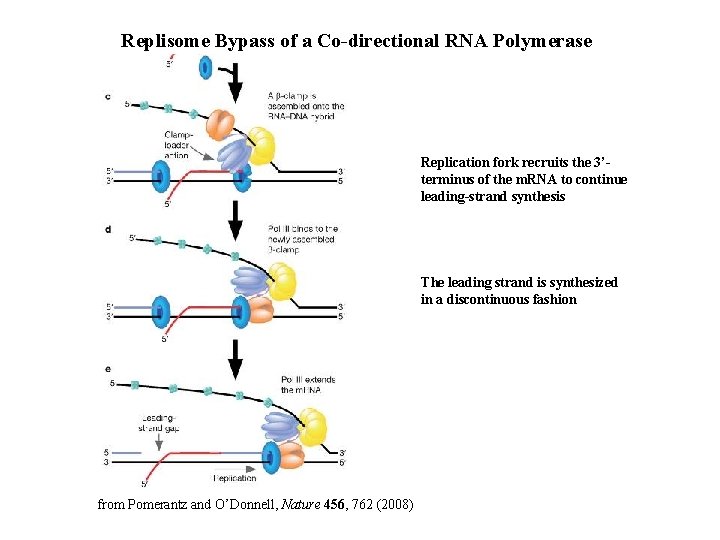
Replisome Bypass of a Co-directional RNA Polymerase Replication fork recruits the 3’terminus of the m. RNA to continue leading-strand synthesis The leading strand is synthesized in a discontinuous fashion from Pomerantz and O’Donnell, Nature 456, 762 (2008)

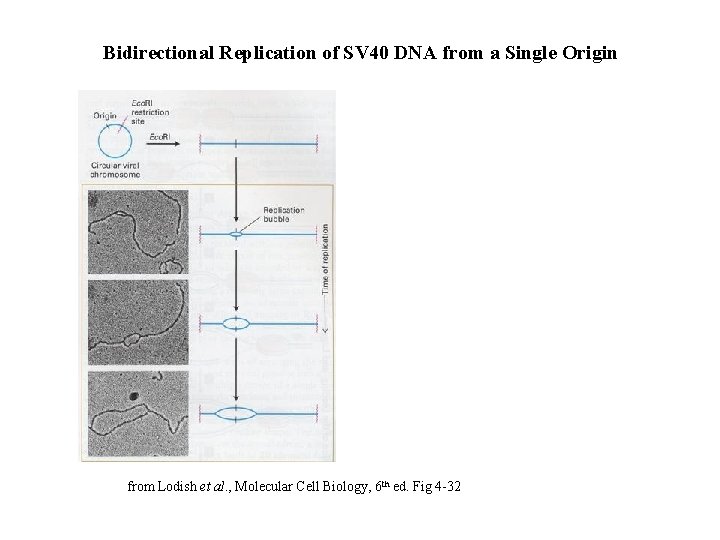
Bidirectional Replication of SV 40 DNA from a Single Origin from Lodish et al. , Molecular Cell Biology, 6 th ed. Fig 4 -32

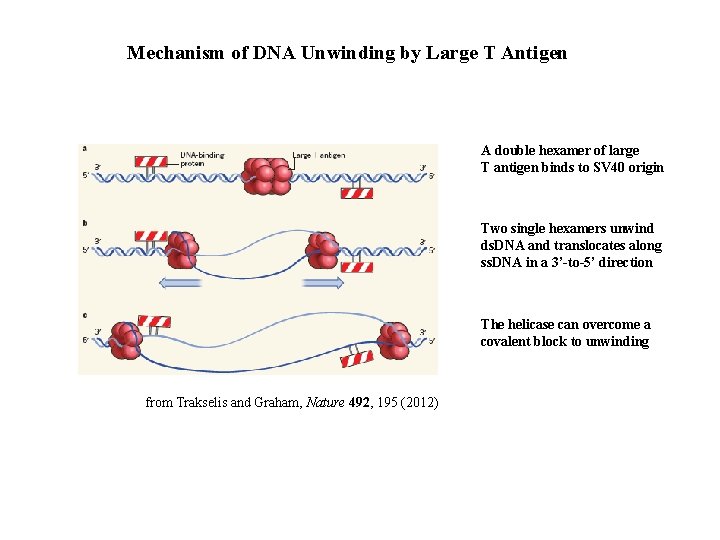
Mechanism of DNA Unwinding by Large T Antigen A double hexamer of large T antigen binds to SV 40 origin Two single hexamers unwind ds. DNA and translocates along ss. DNA in a 3’-to-5’ direction The helicase can overcome a covalent block to unwinding from Trakselis and Graham, Nature 492, 195 (2012)
 Imprinting definition psychology
Imprinting definition psychology Experience expectant vs experience dependent
Experience expectant vs experience dependent Regulacija genske aktivnosti
Regulacija genske aktivnosti Imprinting biology example
Imprinting biology example Genomic imprinting definition
Genomic imprinting definition Dengeli kromozom anomalileri
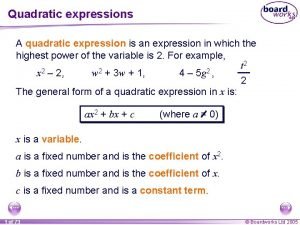
Dengeli kromozom anomalileri Quadratic formula examples
Quadratic formula examples Use the properties of exponents to rewrite each expression
Use the properties of exponents to rewrite each expression Take only photographs leave only footprints
Take only photographs leave only footprints One empire one god one emperor
One empire one god one emperor One one one little dog run
One one one little dog run One king one law one faith
One king one law one faith Byzantine definition
Byzantine definition Ford one plan
Ford one plan See one do one teach one
See one do one teach one See one, do one, teach one
See one, do one, teach one Structure of twelfth night
Structure of twelfth night See one do one teach one
See one do one teach one Asean tourism strategic plan
Asean tourism strategic plan Asean one vision one identity one community
Asean one vision one identity one community Hardy weinberg equation
Hardy weinberg equation Th
Th Allele frequency def
Allele frequency def Allelic frequencies
Allelic frequencies Allele example
Allele example What is a multiple alleles
What is a multiple alleles Calculate allele frequency
Calculate allele frequency Is sickle cell incomplete dominance
Is sickle cell incomplete dominance Codominant allele
Codominant allele When is population in hardy weinberg equilibrium
When is population in hardy weinberg equilibrium Allele example
Allele example Allele vs gene
Allele vs gene Allele vs trait
Allele vs trait