Imaging in Cells via DNA Device Technology DNAPAINT

- Slides: 7

Imaging in Cells via DNA Device Technology: • DNA-PAINT: A Super-Resolution Imaging Technique • HCR FISH • m. RNA Imaging in Cells a. b. c. Ratiometric bimolecular beacons (RBMBs) Multiply labeled tetravalent RNA imaging probes (MTRIPs) Nanoflares 1

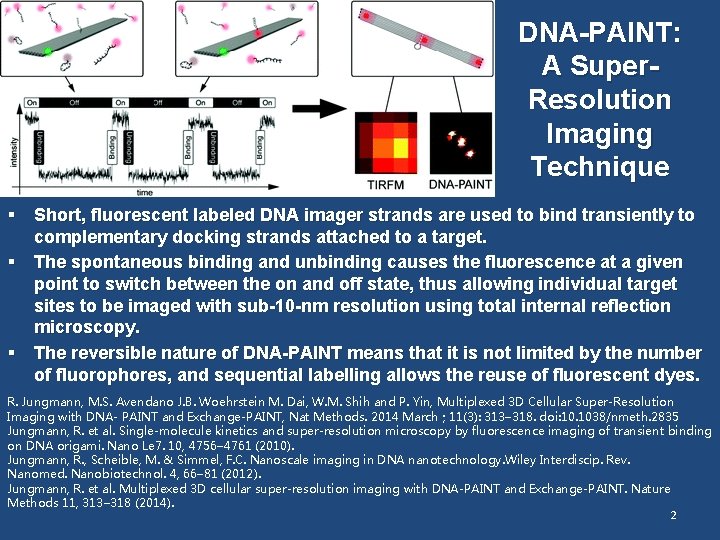
DNA-PAINT: A Super. Resolution Imaging Technique § Short, fluorescent labeled DNA imager strands are used to bind transiently to complementary docking strands attached to a target. § The spontaneous binding and unbinding causes the fluorescence at a given point to switch between the on and off state, thus allowing individual target sites to be imaged with sub-10 -nm resolution using total internal reflection microscopy. § The reversible nature of DNA-PAINT means that it is not limited by the number of fluorophores, and sequential labelling allows the reuse of fluorescent dyes. R. Jungmann, M. S. Avendano J. B. Woehrstein M. Dai, W. M. Shih and P. Yin, Multiplexed 3 D Cellular Super-Resolution Imaging with DNA- PAINT and Exchange-PAINT, Nat Methods. 2014 March ; 11(3): 313– 318. doi: 10. 1038/nmeth. 2835 Jungmann, R. et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Le 7. 10, 4756– 4761 (2010). Jungmann, R. , Scheible, M. & Simmel, F. C. Nanoscale imaging in DNA nanotechnology. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 4, 66– 81 (2012). Jungmann, R. et al. Multiplexed 3 D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nature Methods 11, 313– 318 (2014). 2

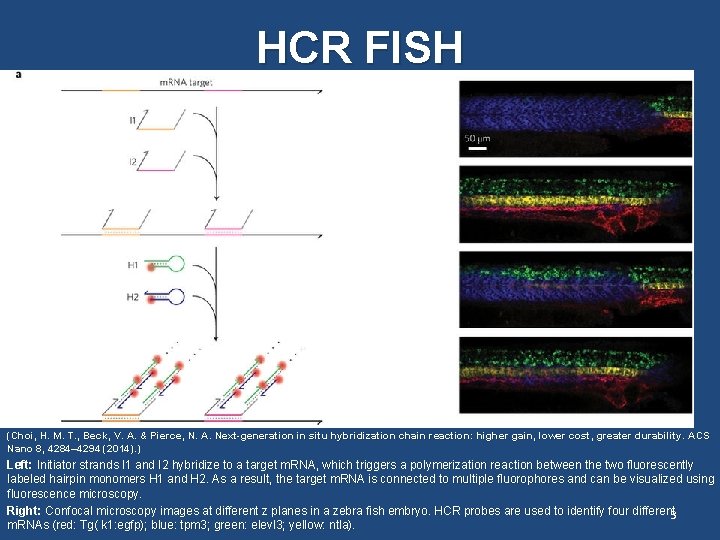
HCR FISH In situ imaging of m. RNA in fixed cells: HCR FISH (Choi, H. M. T. , Beck, V. A. & Pierce, N. A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8, 4284– 4294 (2014). ) Left: Initiator strands I 1 and I 2 hybridize to a target m. RNA, which triggers a polymerization reaction between the two fluorescently labeled hairpin monomers H 1 and H 2. As a result, the target m. RNA is connected to multiple fluorophores and can be visualized using fluorescence microscopy. Right: Confocal microscopy images at different z planes in a zebra fish embryo. HCR probes are used to identify four different 3 m. RNAs (red: Tg( k 1: egfp); blue: tpm 3; green: elevl 3; yellow: ntla).

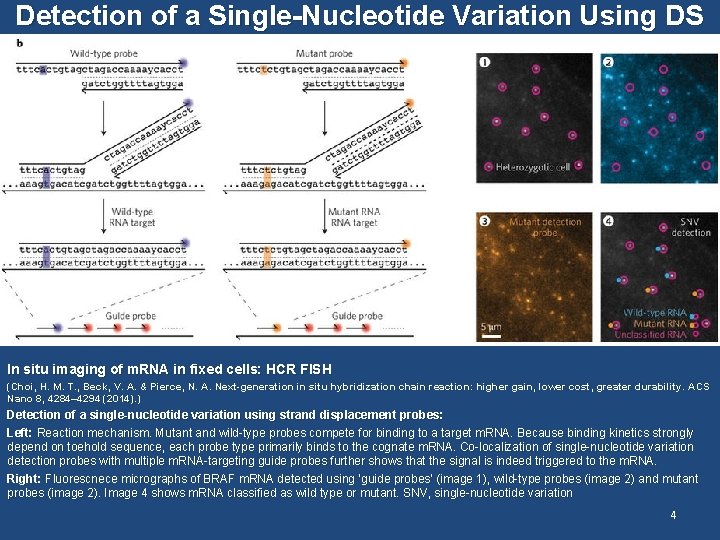
Detection of a Single-Nucleotide Variation Using DS In situ imaging of m. RNA in fixed cells: HCR FISH (Choi, H. M. T. , Beck, V. A. & Pierce, N. A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8, 4284– 4294 (2014). ) Detection of a single-nucleotide variation using strand displacement probes: Left: Reaction mechanism. Mutant and wild-type probes compete for binding to a target m. RNA. Because binding kinetics strongly depend on toehold sequence, each probe type primarily binds to the cognate m. RNA. Co-localization of single-nucleotide variation detection probes with multiple m. RNA-targeting guide probes further shows that the signal is indeed triggered to the m. RNA. Right: Fluorescnece micrographs of BRAF m. RNA detected using ‘guide probes’ (image 1), wild-type probes (image 2) and mutant probes (image 2). Image 4 shows m. RNA classified as wild type or mutant. SNV, single-nucleotide variation 4

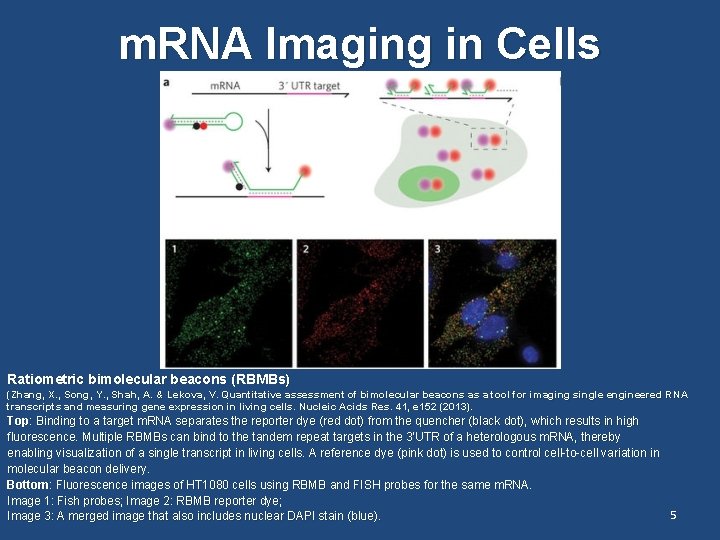
m. RNA Imaging in Cells Ratiometric bimolecular beacons (RBMBs) (Zhang, X. , Song, Y. , Shah, A. & Lekova, V. Quantitative assessment of bimolecular beacons as a tool for imaging single engineered RNA transcripts and measuring gene expression in living cells. Nucleic Acids Res. 41, e 152 (2013). Top: Binding to a target m. RNA separates the reporter dye (red dot) from the quencher (black dot), which results in high fluorescence. Multiple RBMBs can bind to the tandem repeat targets in the 3’UTR of a heterologous m. RNA, thereby enabling visualization of a single transcript in living cells. A reference dye (pink dot) is used to control cell-to-cell variation in molecular beacon delivery. Bottom: Fluorescence images of HT 1080 cells using RBMB and FISH probes for the same m. RNA. Image 1: Fish probes; Image 2: RBMB reporter dye; 5 Image 3: A merged image that also includes nuclear DAPI stain (blue).

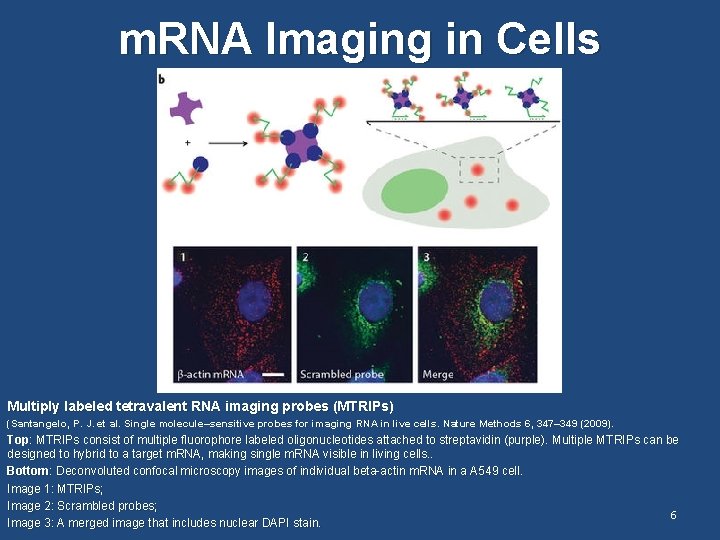
m. RNA Imaging in Cells Multiply labeled tetravalent RNA imaging probes (MTRIPs) (Santangelo, P. J. et al. Single molecule–sensitive probes for imaging RNA in live cells. Nature Methods 6, 347– 349 (2009). Top: MTRIPs consist of multiple fluorophore labeled oligonucleotides attached to streptavidin (purple). Multiple MTRIPs can be designed to hybrid to a target m. RNA, making single m. RNA visible in living cells. . Bottom: Deconvoluted confocal microscopy images of individual beta-actin m. RNA in a A 549 cell. Image 1: MTRIPs; Image 2: Scrambled probes; 6 Image 3: A merged image that includes nuclear DAPI stain.

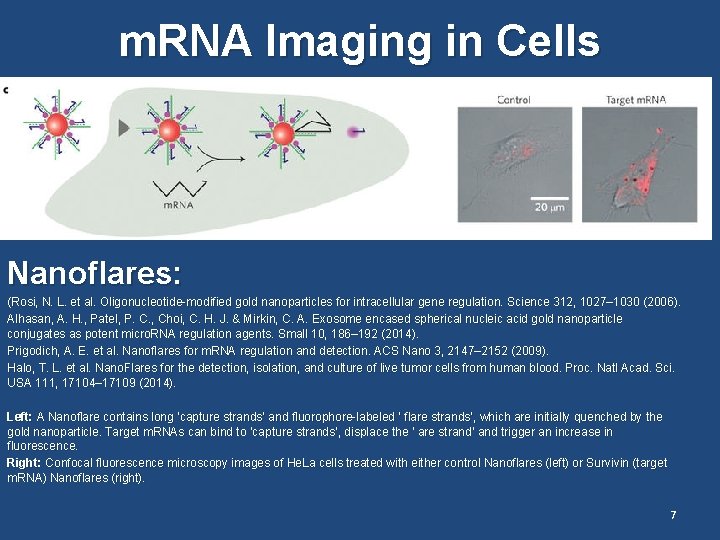
m. RNA Imaging in Cells Nanoflares: (Rosi, N. L. et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 312, 1027– 1030 (2006). Alhasan, A. H. , Patel, P. C. , Choi, C. H. J. & Mirkin, C. A. Exosome encased spherical nucleic acid gold nanoparticle conjugates as potent micro. RNA regulation agents. Small 10, 186– 192 (2014). Prigodich, A. E. et al. Nanoflares for m. RNA regulation and detection. ACS Nano 3, 2147– 2152 (2009). Halo, T. L. et al. Nano. Flares for the detection, isolation, and culture of live tumor cells from human blood. Proc. Natl Acad. Sci. USA 111, 17104– 17109 (2014). Left: A Nanoflare contains long ‘capture strands’ and fluorophore-labeled ‘ flare strands’, which are initially quenched by the gold nanoparticle. Target m. RNAs can bind to ‘capture strands’, displace the ‘ are strand’ and trigger an increase in fluorescence. Right: Confocal fluorescence microscopy images of He. La cells treated with either control Nanoflares (left) or Survivin (target m. RNA) Nanoflares (right). 7