Identification of structurally diverse Growth Hormone Secretagogue GHS

- Slides: 10

Identification of structurally diverse Growth Hormone Secretagogue (GHS) agonists by virtual screening and structure-activity relationship analysis of 2 -formylaminoacetamide derivatives. J. Med. Chem. 2004, 47, 4286 -4290 GHS controls the release of growth hormone from the pituitary gland Therapeutically, GHS agonists replace direct growth hormone administration - growth deficiency - osteoporosis - obesity

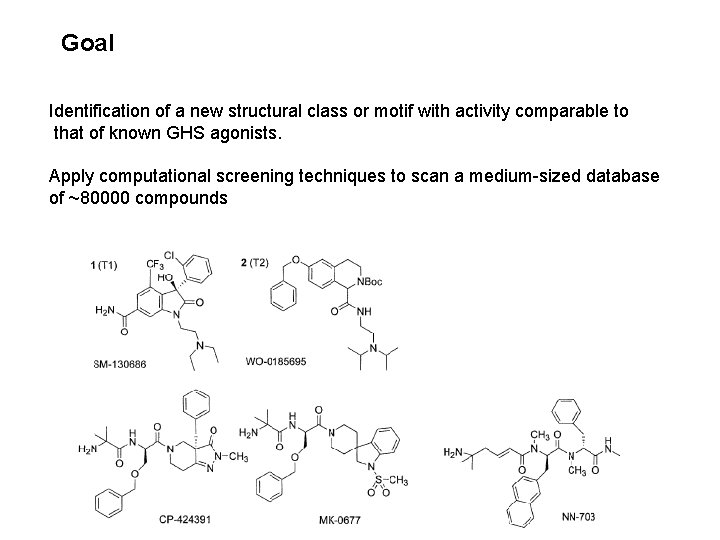
Goal Identification of a new structural class or motif with activity comparable to that of known GHS agonists. Apply computational screening techniques to scan a medium-sized database of ~80000 compounds

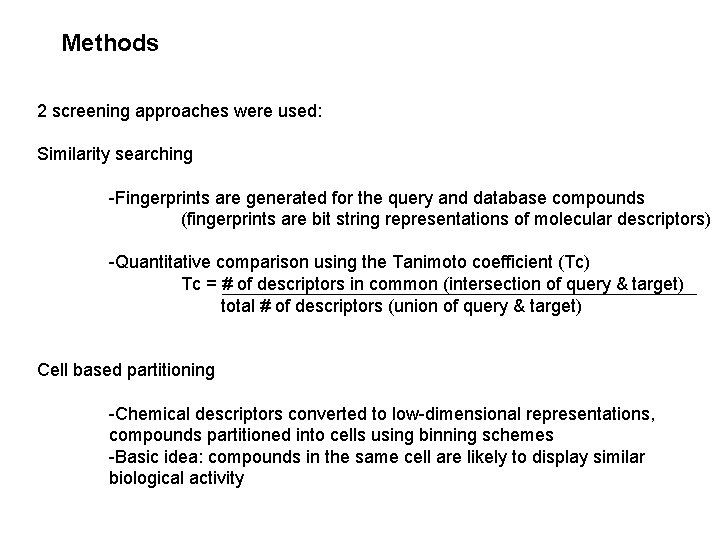
Methods 2 screening approaches were used: Similarity searching -Fingerprints are generated for the query and database compounds (fingerprints are bit string representations of molecular descriptors) -Quantitative comparison using the Tanimoto coefficient (Tc) Tc = # of descriptors in common (intersection of query & target) total # of descriptors (union of query & target) Cell based partitioning -Chemical descriptors converted to low-dimensional representations, compounds partitioned into cells using binning schemes -Basic idea: compounds in the same cell are likely to display similar biological activity

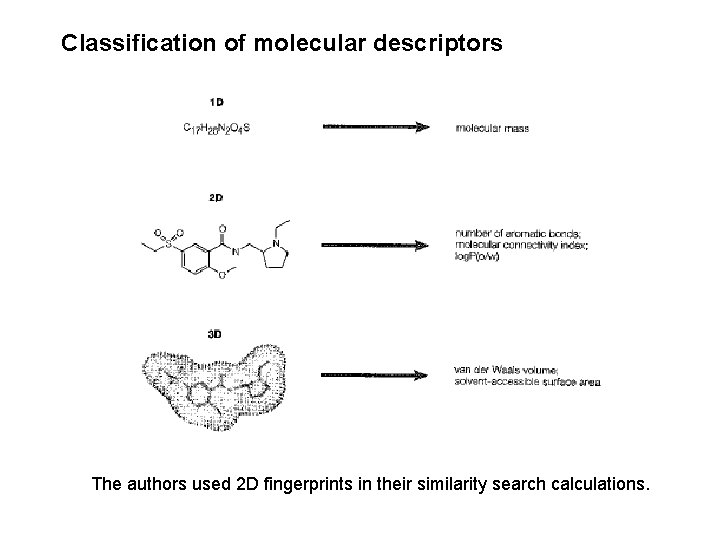
Classification of molecular descriptors The authors used 2 D fingerprints in their similarity search calculations.

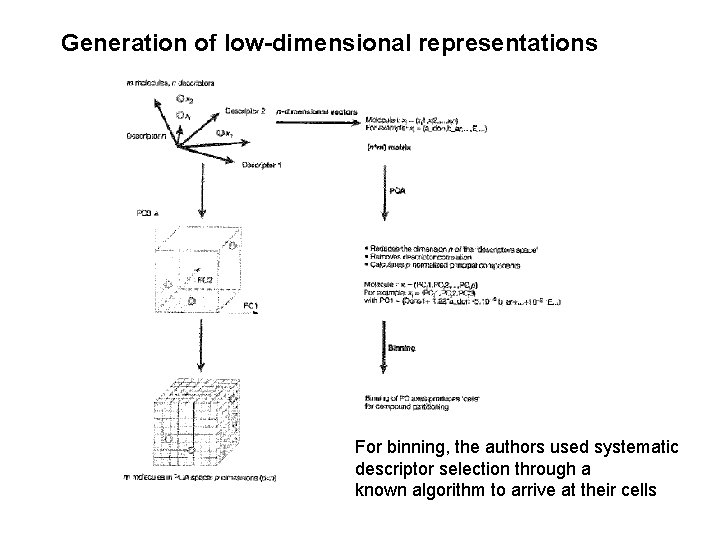
Generation of low-dimensional representations For binning, the authors used systematic descriptor selection through a known algorithm to arrive at their cells

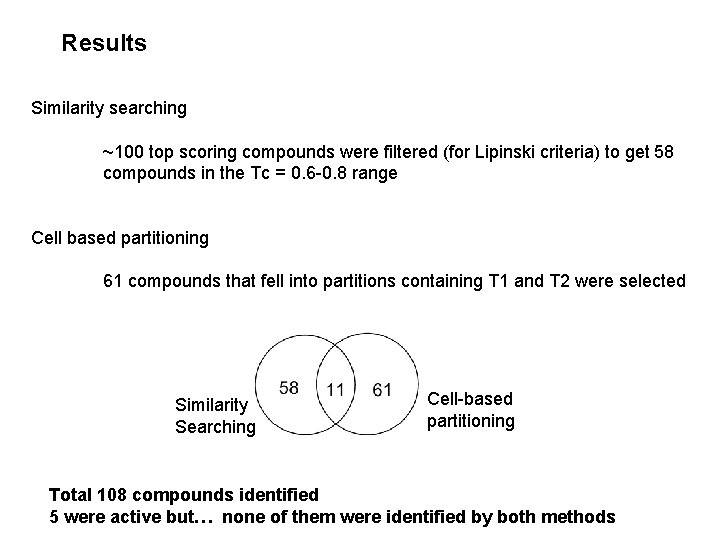
Results Similarity searching ~100 top scoring compounds were filtered (for Lipinski criteria) to get 58 compounds in the Tc = 0. 6 -0. 8 range Cell based partitioning 61 compounds that fell into partitions containing T 1 and T 2 were selected Similarity Searching Cell-based partitioning Total 108 compounds identified 5 were active but… none of them were identified by both methods

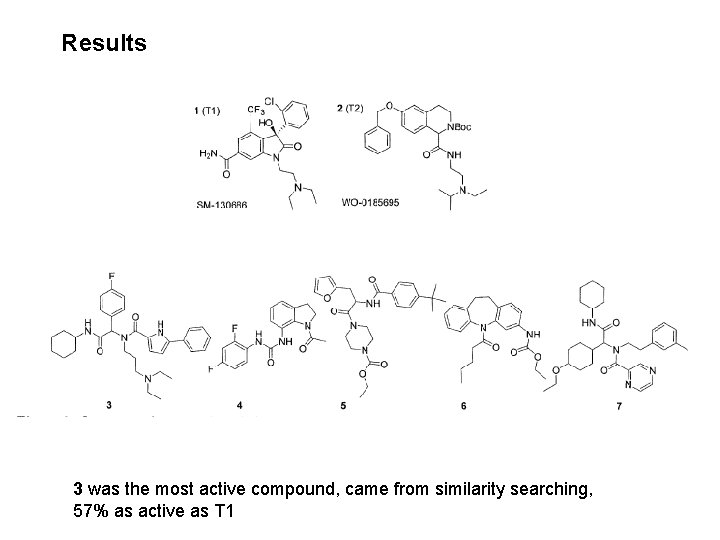
Results 3 was the most active compound, came from similarity searching, 57% as active as T 1

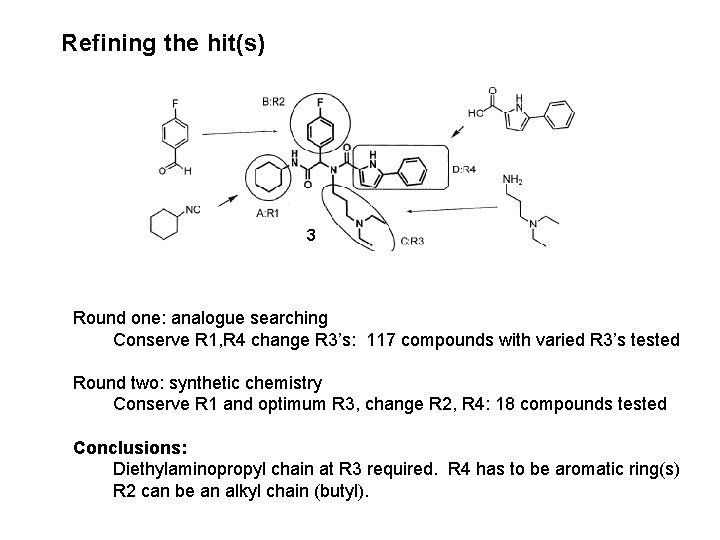
Refining the hit(s) 3 Round one: analogue searching Conserve R 1, R 4 change R 3’s: 117 compounds with varied R 3’s tested Round two: synthetic chemistry Conserve R 1 and optimum R 3, change R 2, R 4: 18 compounds tested Conclusions: Diethylaminopropyl chain at R 3 required. R 4 has to be aromatic ring(s) R 2 can be an alkyl chain (butyl).

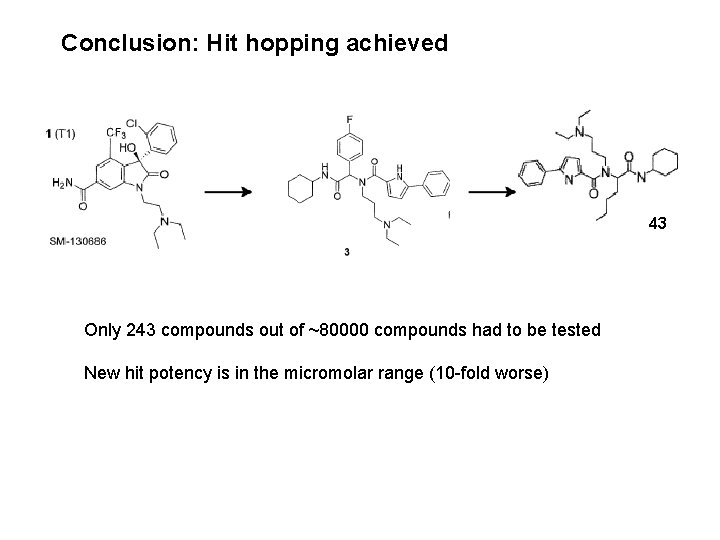
Conclusion: Hit hopping achieved 43 Only 243 compounds out of ~80000 compounds had to be tested New hit potency is in the micromolar range (10 -fold worse)

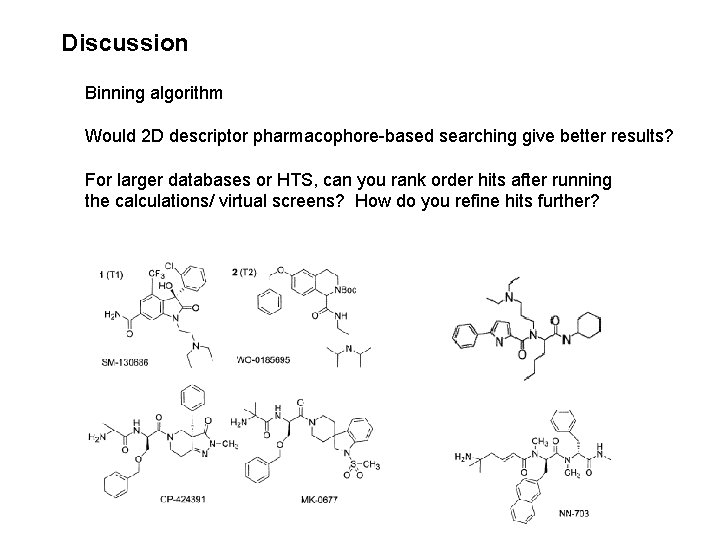
Discussion Binning algorithm Would 2 D descriptor pharmacophore-based searching give better results? For larger databases or HTS, can you rank order hits after running the calculations/ virtual screens? How do you refine hits further?