Ideas for Integrating Campus Resources with Galaxy Framework


- Slides: 12

Ideas for Integrating Campus Resources with Galaxy Framework Ravi K Madduri Argonne National Laboratory Computation Institute, University of Chicago @madduri www. ci. anl. gov www. ci. uchicago. edu

Outline Challenges in Sequencing Analysis • Our approach using AWS • Benefits of using Galaxy with Globus Online • Galaxy architecture on cloud • – Enabling HTC through Condor and Swift Ongoing engagements and Lessons learned • Leveraging Campus infrastructure • Challenges – Potential solutions – 2 www. ci. anl. gov www. ci. uchicago. edu

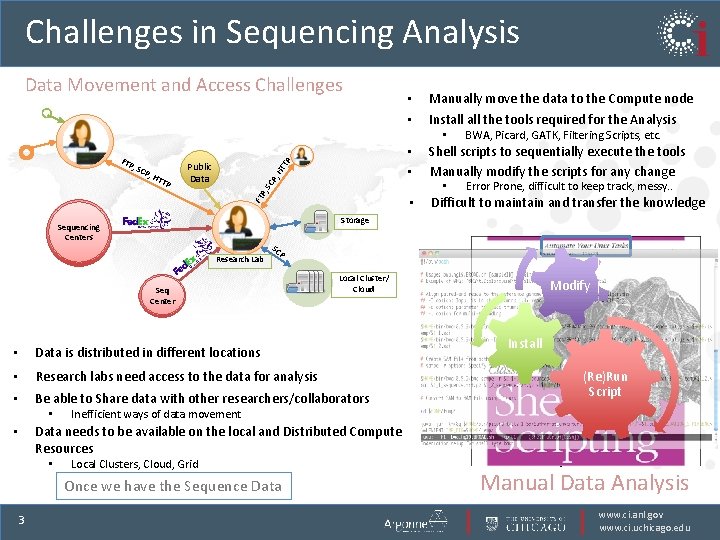
Challenges in Sequencing Analysis Data Movement and Access Challenges • • Manually move the data to the Compute node Install the tools required for the Analysis • TTP TP Public Data HT P, H SC P, , SC • • FT • Research Lab P Local Cluster/ Cloud • Data is distributed in different locations • Research labs need access to the data for analysis • Be able to Share data with other researchers/collaborators Inefficient ways of data movement Data needs to be available on the local and Distributed Compute Resources • Local Clusters, Cloud, Grid Once we have the Sequence Data 3 Difficult to maintain and transfer the knowledge Fastq SC Seq Center • Error Prone, difficult to keep track, messy. . Storage Sequencing Centers • Shell scripts to sequentially execute the tools Manually modify the scripts for any change • P, FTP BWA, Picard, GATK, Filtering Scripts, etc. Ref Genome Modify Picard Install Alignment (Re)Run GATK Script Variant Calling How do we analyze this Sequence Data Manual Data Analysis www. ci. anl. gov www. ci. uchicago. edu

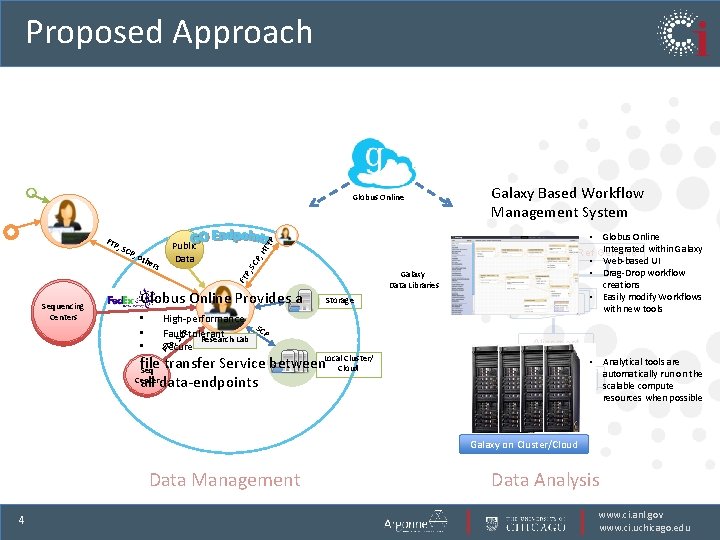
Proposed Approach the r Fastq Galaxy Data Libraries FT P, s TT P, o Public Data , H , SC SC P FTP Sequencing Centers Globus Online Provides a • • • Galaxy Based Workflow Management System P Globus Online High-performance SC P Fault-tolerant CP Research Lab S , P Secure FT Storage Cluster/ file transfer Service between. Local. Cloud Seq Center all data-endpoints Picard • Globus Online Integrated within Galaxy Ref Genome • Web-based UI • Drag-Drop workflow creations • Easily modify Workflows with new tools Alignment GATK • Analytical tools are automatically run on the scalable compute resources when possible Variant Calling Galaxy on Cluster/Cloud Data Management 4 Data Analysis www. ci. anl. gov www. ci. uchicago. edu

Benefits of Globus Online • Globus Online - No IT required – Software as a Service (Saa. S) • • No client software installation New features automatically available – Consolidated support & troubleshooting – Works with existing Grid. FTP servers – Globus Connect solves “last mile problem” • Grid. FTP-based – High-performance, reliable data transfer protocol optimized for high-bandwidth wide-area networks – Low bandwidth; encrypted and integrity protected by default – Defacto standard for data movement in large national cyberinfrastructure projects • >8000 registered users, >10 PB moved • Recommended and used by DOE Facilities, NSF Supercomputing centers, and many campuses 5 www. ci. anl. gov www. ci. uchicago. edu

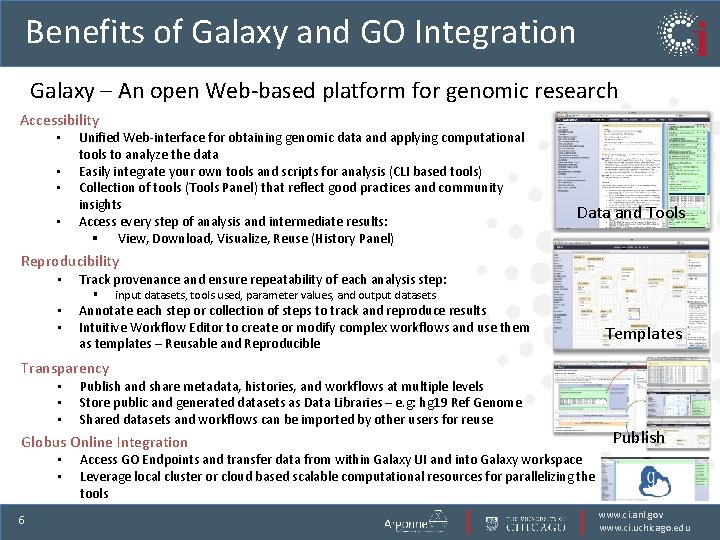
Benefits of Galaxy and GO Integration Galaxy – An open Web-based platform for genomic research Accessibility • • Unified Web-interface for obtaining genomic data and applying computational tools to analyze the data Easily integrate your own tools and scripts for analysis (CLI based tools) Collection of tools (Tools Panel) that reflect good practices and community insights Access every step of analysis and intermediate results: § View, Download, Visualize, Reuse (History Panel) Data and Tools Reproducibility • Track provenance and ensure repeatability of each analysis step: § • • input datasets, tools used, parameter values, and output datasets Annotate each step or collection of steps to track and reproduce results Intuitive Workflow Editor to create or modify complex workflows and use them as templates – Reusable and Reproducible Templates Transparency • • • Publish and share metadata, histories, and workflows at multiple levels Store public and generated datasets as Data Libraries – e. g: hg 19 Ref Genome Shared datasets and workflows can be imported by other users for reuse Globus Online Integration • • 6 Publish Access GO Endpoints and transfer data from within Galaxy UI and into Galaxy workspace Leverage local cluster or cloud based scalable computational resources for parallelizing the tools www. ci. anl. gov www. ci. uchicago. edu

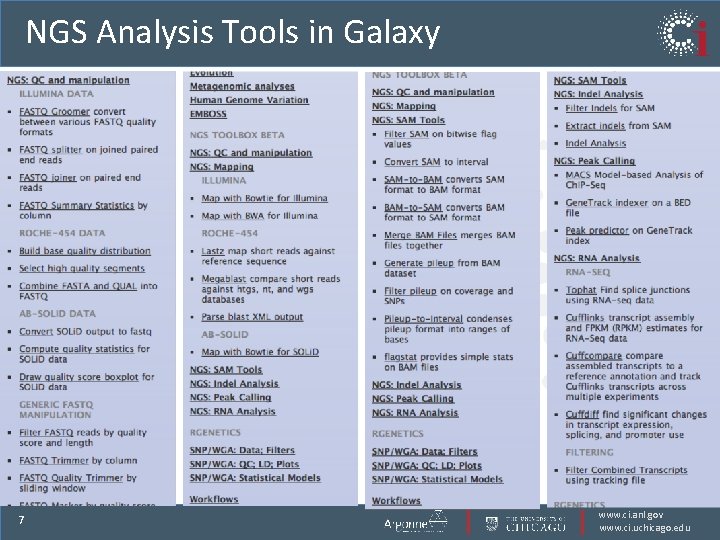
NGS Analysis Tools in Galaxy 7 www. ci. anl. gov www. ci. uchicago. edu

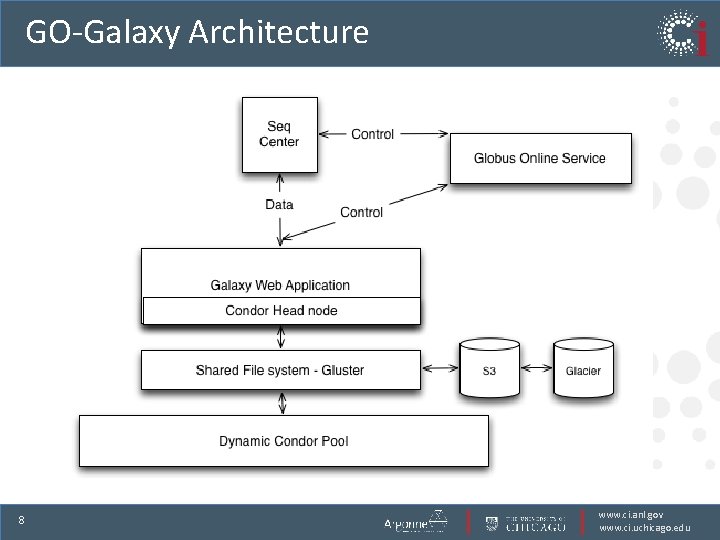
GO-Galaxy Architecture 8 www. ci. anl. gov www. ci. uchicago. edu

Using Galaxy for other domains Galaxy for Cosmology – PDACS • Galaxy for Climate modeling – FACE-IT • Galaxy for Light sources • 9 www. ci. anl. gov www. ci. uchicago. edu

Integrating Campus resources • Issues Galaxy uses a shared file system where worker nodes look for input files and store output files – Installing Applications/Executables and dependencies – Location of Reference datasets – 10 www. ci. anl. gov www. ci. uchicago. edu

Integrating Campus Resources Using LWR created by John Chilton • Staging the data in to compute nodes • Condor-G – Swift • Using Parrot (http: //nd. edu/~ccl/software/parrot/) to intercept disk I/O • Using Object store support – 11 www. ci. anl. gov www. ci. uchicago. edu

Acknowledgements Ideas largely from discussions with Jamie Frey, Mike Wilde, Suchandra Thapa, John Chilton • NHLBI-funded CVRG project • Amazon Research Grants • Globus Online team at the CI/ANL • 12 www. ci. anl. gov www. ci. uchicago. edu