IBD sharing in the 1000 Genomes Project Phase

IBD sharing in the 1000 Genomes Project Phase 3 data reveals relationships from Neandertals to present day families Gundula Povysil Johannes Kepler University Linz Austria
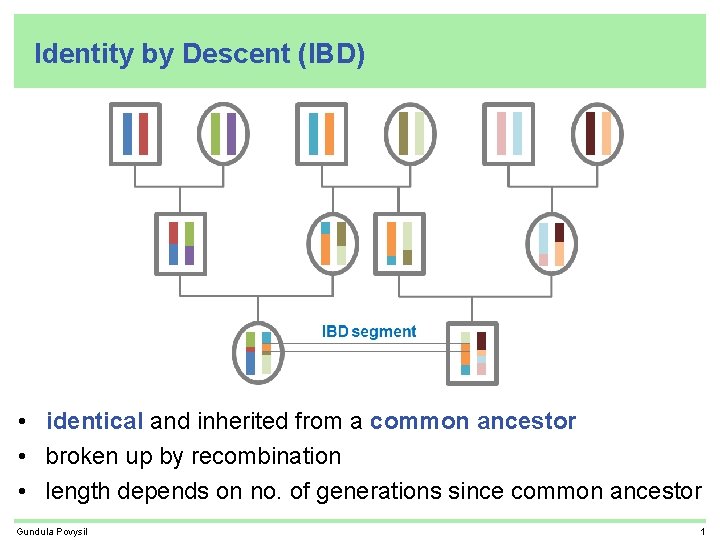
Identity by Descent (IBD) • identical and inherited from a common ancestor • broken up by recombination • length depends on no. of generations since common ancestor Gundula Povysil 1

Identity by Descent (IBD) • identical and inherited from a common ancestor • broken up by recombination • length depends on no. of generations since common ancestor Gundula Povysil 1
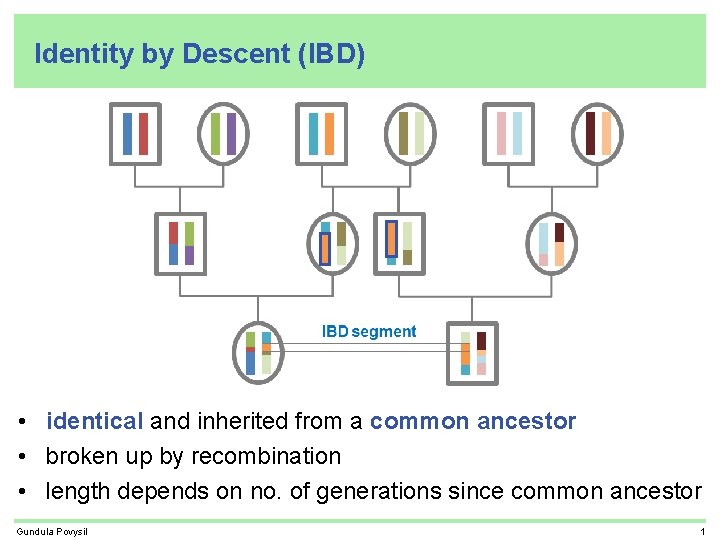
Identity by Descent (IBD) • identical and inherited from a common ancestor • broken up by recombination • length depends on no. of generations since common ancestor Gundula Povysil 1
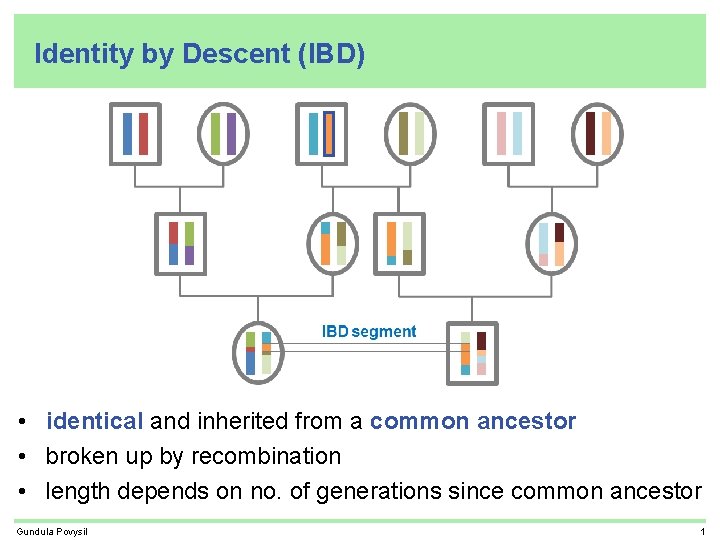
Identity by Descent (IBD) • identical and inherited from a common ancestor • broken up by recombination • length depends on no. of generations since common ancestor Gundula Povysil 1
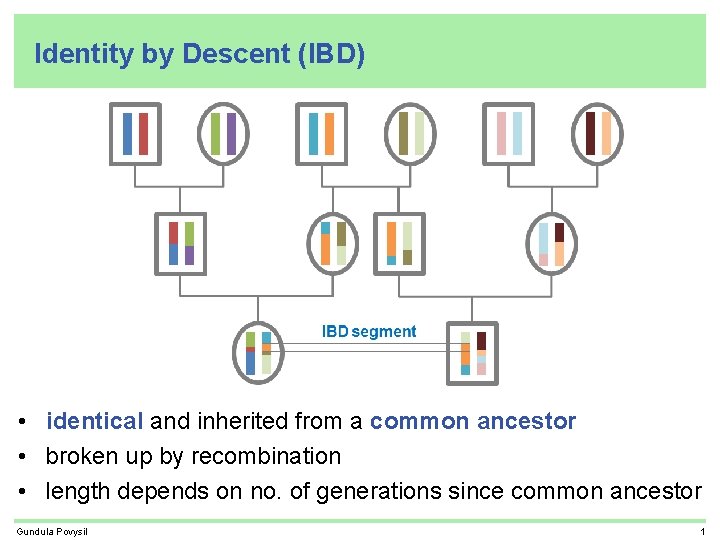
Identity by Descent (IBD) • identical and inherited from a common ancestor • broken up by recombination • length depends on no. of generations since common ancestor Gundula Povysil 1
Hap. FABIA (Hochreiter 2013) • biclustering of SNV data to find short IBD segments shared by multiple individuals • based on low-frequency and rare SNVs (MAF < 5%) Gundula Povysil 2
Hap. FABIA (Hochreiter 2013) • biclustering of SNV data to find short IBD segments shared by multiple individuals • based on low-frequency and rare SNVs (MAF < 5%) Steps: 1. biclustering algorithm extracts subsets of individuals that share a subset of SNVs 2. Hap. FABIA extracts IBD segments as local accumulations of shared SNVs (tag. SNVs) from biclusters Gundula Povysil 2
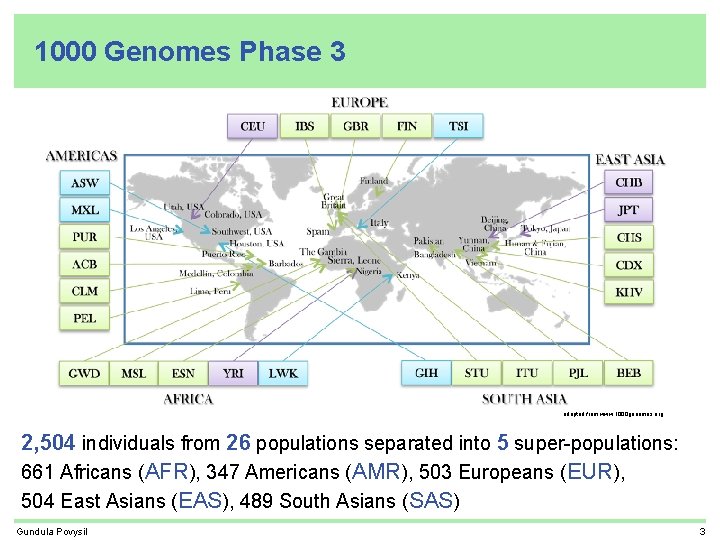
1000 Genomes Phase 3 adapted from www. 1000 genomes. org 2, 504 individuals from 26 populations separated into 5 super-populations: 661 Africans (AFR), 347 Americans (AMR), 503 Europeans (EUR), 504 East Asians (EAS), 489 South Asians (SAS) Gundula Povysil 3
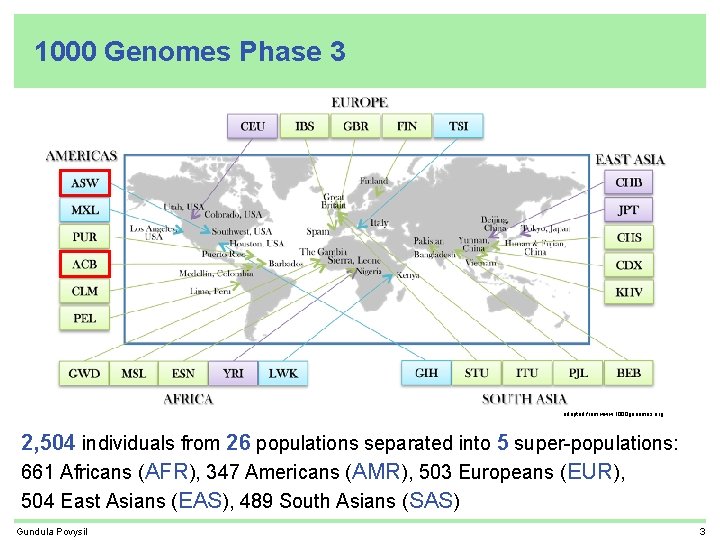
1000 Genomes Phase 3 adapted from www. 1000 genomes. org 2, 504 individuals from 26 populations separated into 5 super-populations: 661 Africans (AFR), 347 Americans (AMR), 503 Europeans (EUR), 504 East Asians (EAS), 489 South Asians (SAS) Gundula Povysil 3
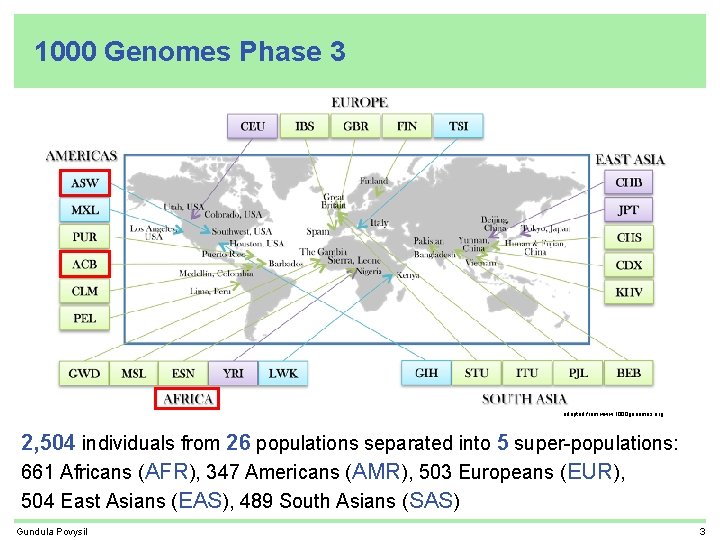
1000 Genomes Phase 3 adapted from www. 1000 genomes. org 2, 504 individuals from 26 populations separated into 5 super-populations: 661 Africans (AFR), 347 Americans (AMR), 503 Europeans (EUR), 504 East Asians (EAS), 489 South Asians (SAS) Gundula Povysil 3
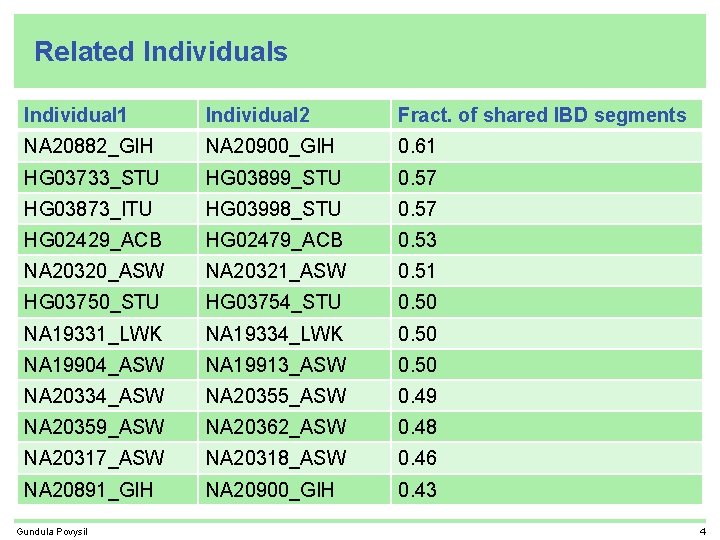
Related Individuals Individual 1 Individual 2 Fract. of shared IBD segments NA 20882_GIH NA 20900_GIH 0. 61 HG 03733_STU HG 03899_STU 0. 57 HG 03873_ITU HG 03998_STU 0. 57 HG 02429_ACB HG 02479_ACB 0. 53 NA 20320_ASW NA 20321_ASW 0. 51 HG 03750_STU HG 03754_STU 0. 50 NA 19331_LWK NA 19334_LWK 0. 50 NA 19904_ASW NA 19913_ASW 0. 50 NA 20334_ASW NA 20355_ASW 0. 49 NA 20359_ASW NA 20362_ASW 0. 48 NA 20317_ASW NA 20318_ASW 0. 46 NA 20891_GIH NA 20900_GIH 0. 43 Gundula Povysil 4
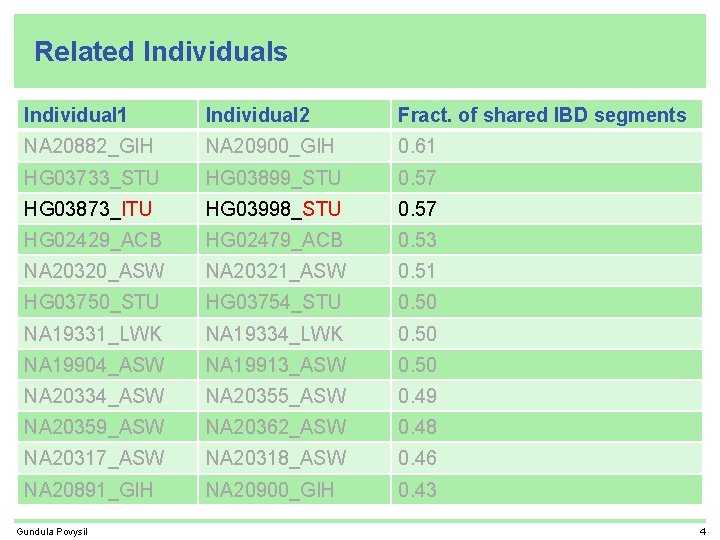
Related Individuals Individual 1 Individual 2 Fract. of shared IBD segments NA 20882_GIH NA 20900_GIH 0. 61 HG 03733_STU HG 03899_STU 0. 57 HG 03873_ITU HG 03998_STU 0. 57 HG 02429_ACB HG 02479_ACB 0. 53 NA 20320_ASW NA 20321_ASW 0. 51 HG 03750_STU HG 03754_STU 0. 50 NA 19331_LWK NA 19334_LWK 0. 50 NA 19904_ASW NA 19913_ASW 0. 50 NA 20334_ASW NA 20355_ASW 0. 49 NA 20359_ASW NA 20362_ASW 0. 48 NA 20317_ASW NA 20318_ASW 0. 46 NA 20891_GIH NA 20900_GIH 0. 43 Gundula Povysil 4
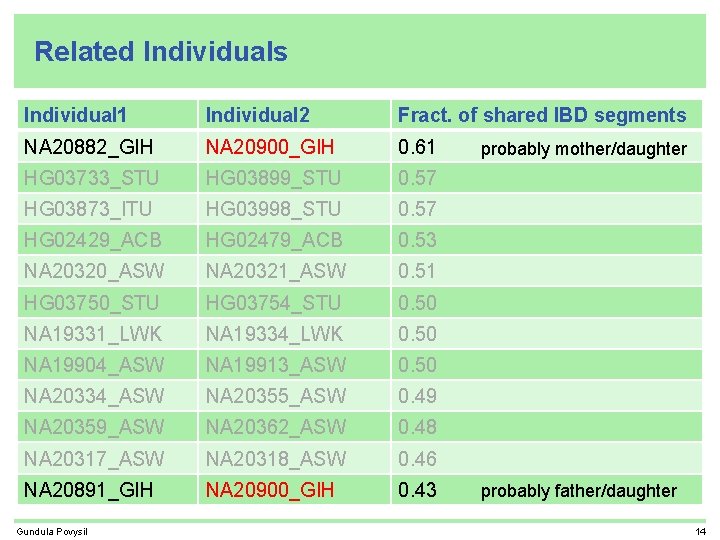
Related Individuals Individual 1 Individual 2 Fract. of shared IBD segments NA 20882_GIH NA 20900_GIH 0. 61 HG 03733_STU HG 03899_STU 0. 57 HG 03873_ITU HG 03998_STU 0. 57 HG 02429_ACB HG 02479_ACB 0. 53 NA 20320_ASW NA 20321_ASW 0. 51 HG 03750_STU HG 03754_STU 0. 50 NA 19331_LWK NA 19334_LWK 0. 50 NA 19904_ASW NA 19913_ASW 0. 50 NA 20334_ASW NA 20355_ASW 0. 49 NA 20359_ASW NA 20362_ASW 0. 48 NA 20317_ASW NA 20318_ASW 0. 46 NA 20891_GIH NA 20900_GIH 0. 43 Gundula Povysil probably mother/daughter probably father/daughter 14
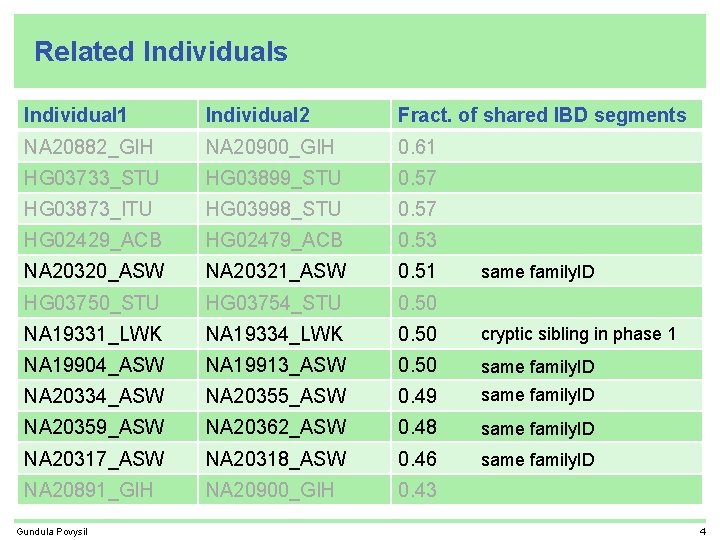
Related Individuals Individual 1 Individual 2 Fract. of shared IBD segments NA 20882_GIH NA 20900_GIH 0. 61 HG 03733_STU HG 03899_STU 0. 57 HG 03873_ITU HG 03998_STU 0. 57 HG 02429_ACB HG 02479_ACB 0. 53 NA 20320_ASW NA 20321_ASW 0. 51 HG 03750_STU HG 03754_STU 0. 50 NA 19331_LWK NA 19334_LWK 0. 50 cryptic sibling in phase 1 NA 19904_ASW NA 19913_ASW 0. 50 same family. ID NA 20334_ASW NA 20355_ASW 0. 49 same family. ID NA 20359_ASW NA 20362_ASW 0. 48 same family. ID NA 20317_ASW NA 20318_ASW 0. 46 same family. ID NA 20891_GIH NA 20900_GIH 0. 43 Gundula Povysil same family. ID 4
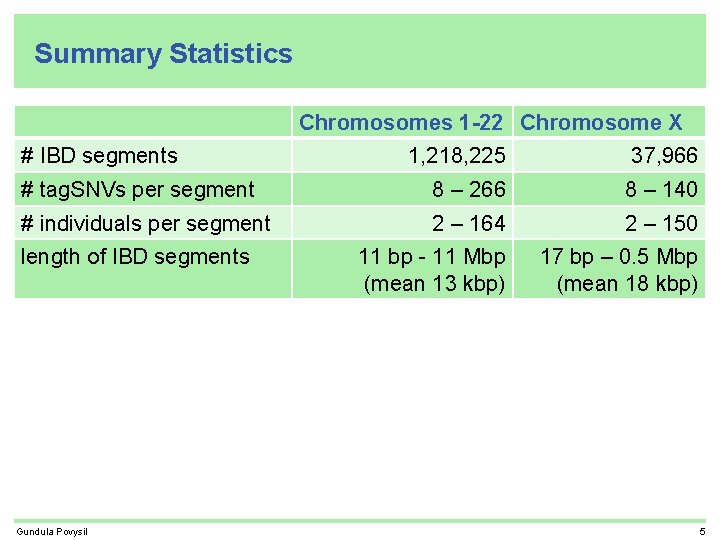
Summary Statistics Chromosomes 1 -22 Chromosome X # IBD segments 1, 218, 225 37, 966 # tag. SNVs per segment 8 – 266 8 – 140 # individuals per segment 2 – 164 2 – 150 11 bp - 11 Mbp (mean 13 kbp) 17 bp – 0. 5 Mbp (mean 18 kbp) length of IBD segments Gundula Povysil 5
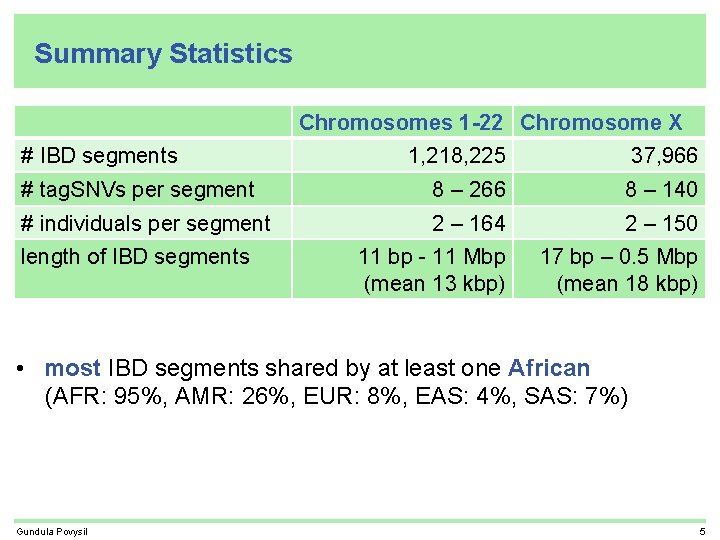
Summary Statistics Chromosomes 1 -22 Chromosome X # IBD segments 1, 218, 225 37, 966 # tag. SNVs per segment 8 – 266 8 – 140 # individuals per segment 2 – 164 2 – 150 11 bp - 11 Mbp (mean 13 kbp) 17 bp – 0. 5 Mbp (mean 18 kbp) length of IBD segments • most IBD segments shared by at least one African (AFR: 95%, AMR: 26%, EUR: 8%, EAS: 4%, SAS: 7%) Gundula Povysil 5
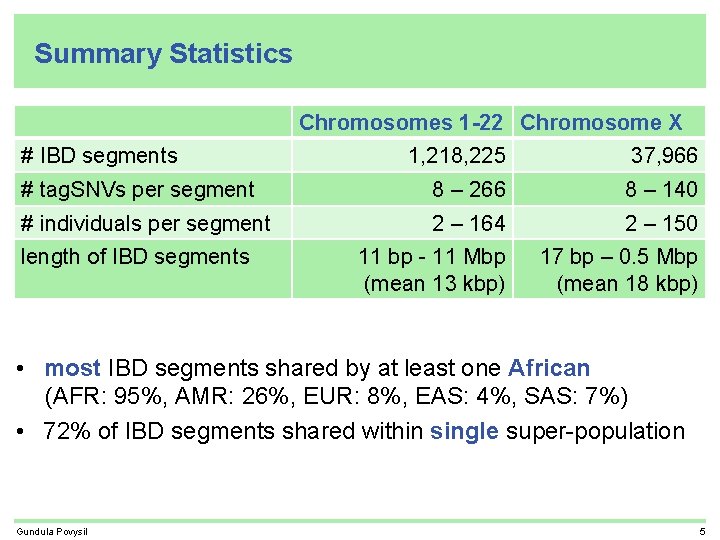
Summary Statistics Chromosomes 1 -22 Chromosome X # IBD segments 1, 218, 225 37, 966 # tag. SNVs per segment 8 – 266 8 – 140 # individuals per segment 2 – 164 2 – 150 11 bp - 11 Mbp (mean 13 kbp) 17 bp – 0. 5 Mbp (mean 18 kbp) length of IBD segments • most IBD segments shared by at least one African (AFR: 95%, AMR: 26%, EUR: 8%, EAS: 4%, SAS: 7%) • 72% of IBD segments shared within single super-population Gundula Povysil 5
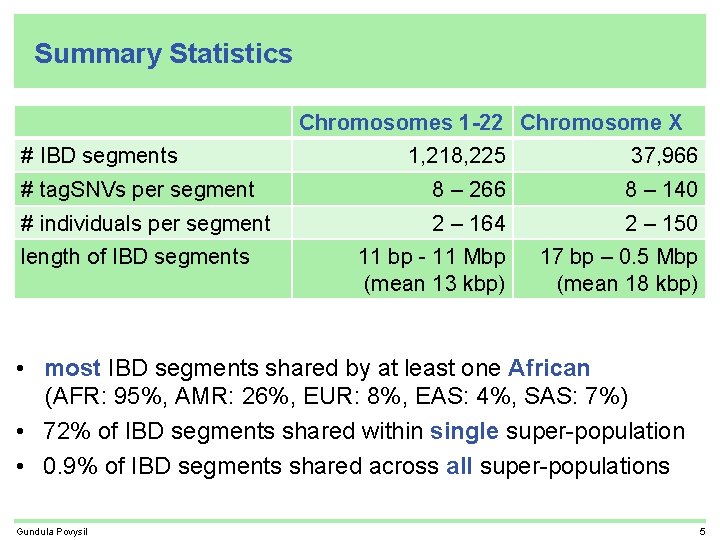
Summary Statistics Chromosomes 1 -22 Chromosome X # IBD segments 1, 218, 225 37, 966 # tag. SNVs per segment 8 – 266 8 – 140 # individuals per segment 2 – 164 2 – 150 11 bp - 11 Mbp (mean 13 kbp) 17 bp – 0. 5 Mbp (mean 18 kbp) length of IBD segments • most IBD segments shared by at least one African (AFR: 95%, AMR: 26%, EUR: 8%, EAS: 4%, SAS: 7%) • 72% of IBD segments shared within single super-population • 0. 9% of IBD segments shared across all super-populations Gundula Povysil 5
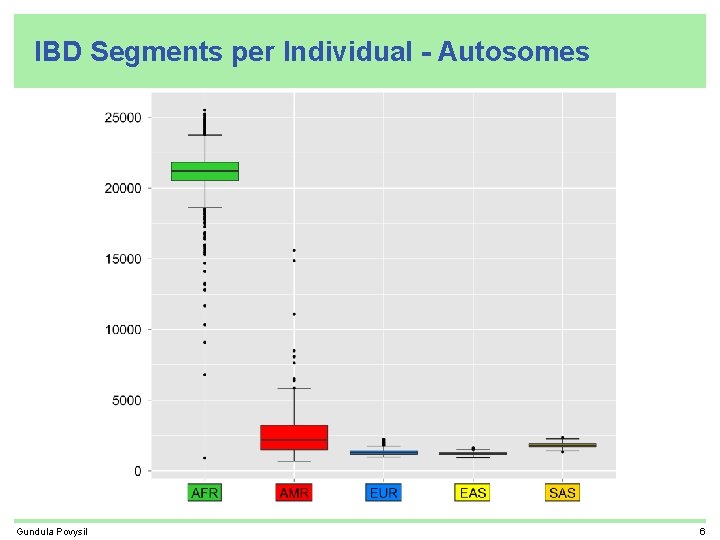
IBD Segments per Individual - Autosomes Gundula Povysil 6
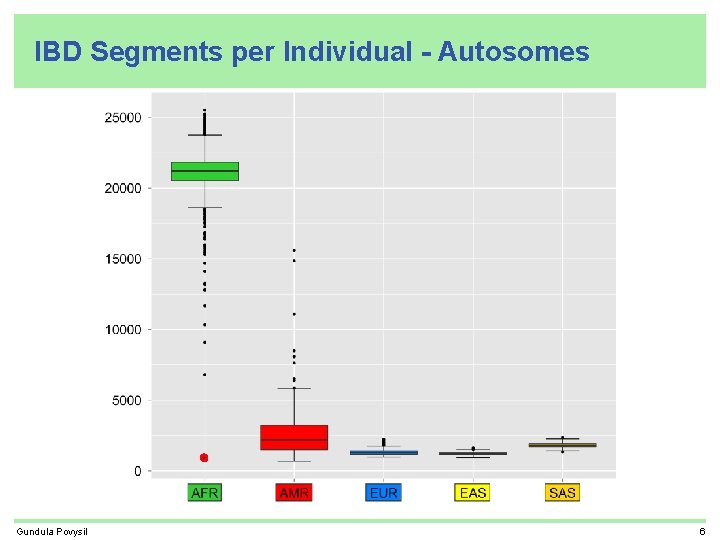
IBD Segments per Individual - Autosomes Gundula Povysil 6
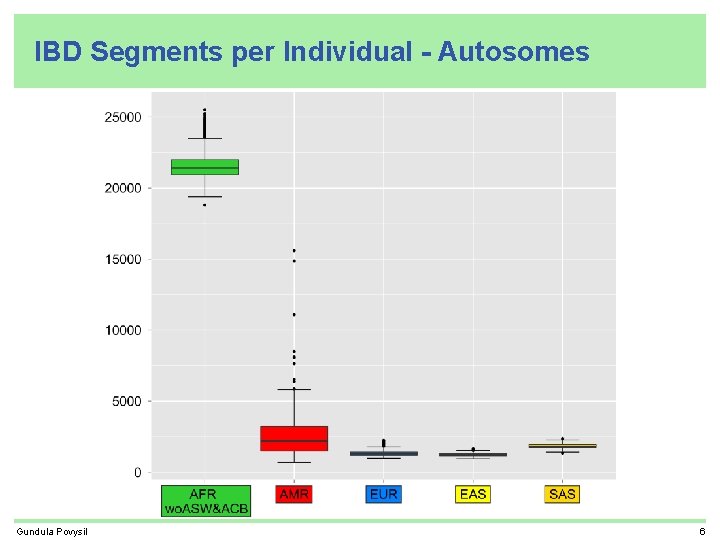
IBD Segments per Individual - Autosomes Gundula Povysil 6
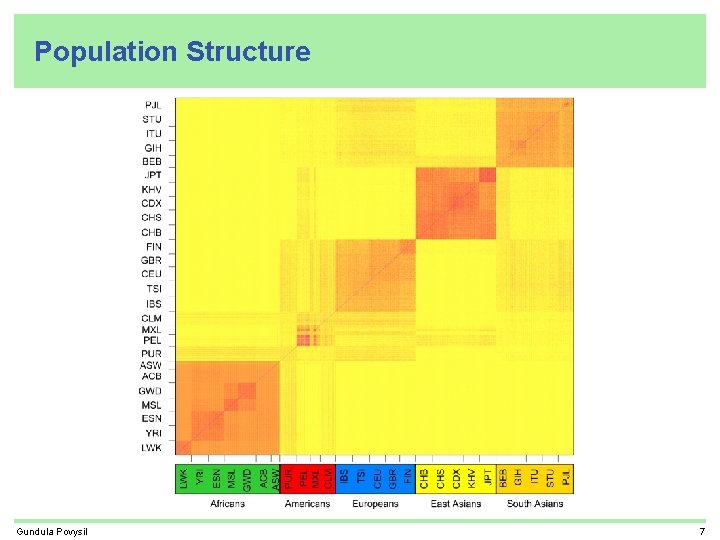
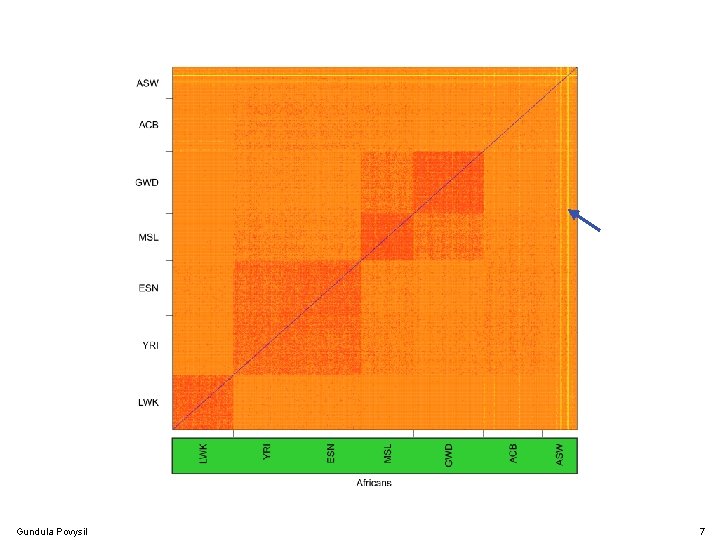
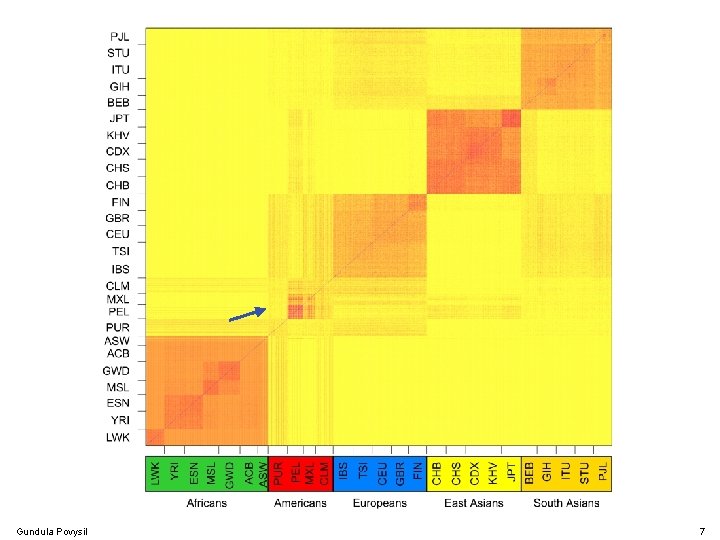
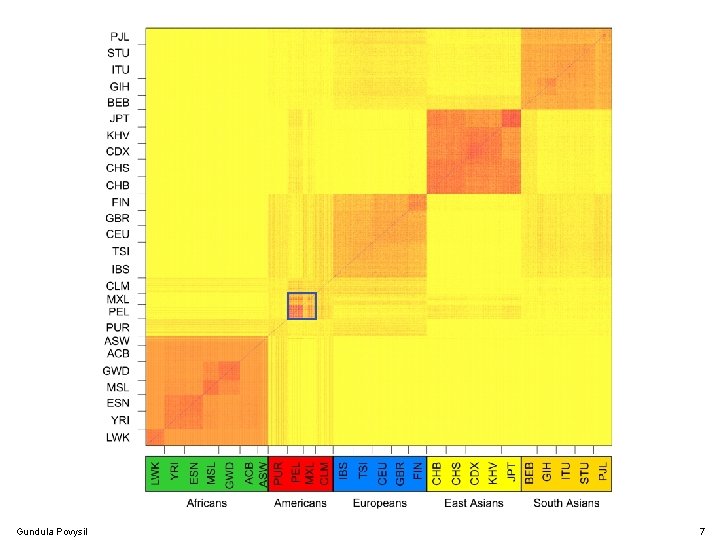
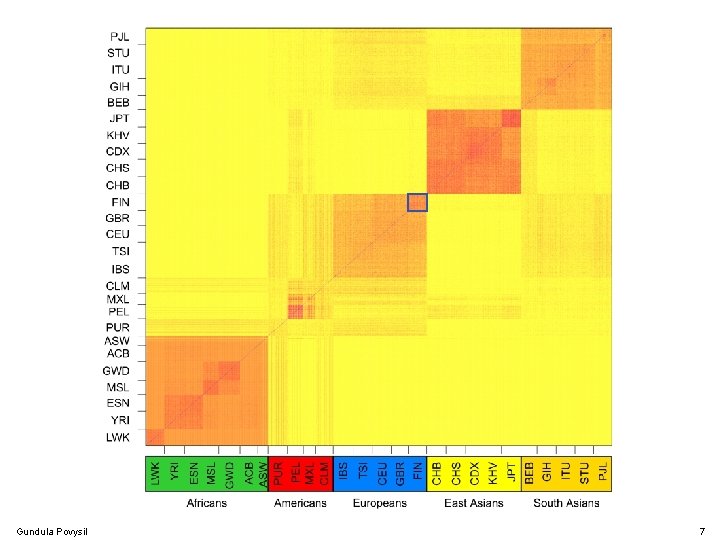
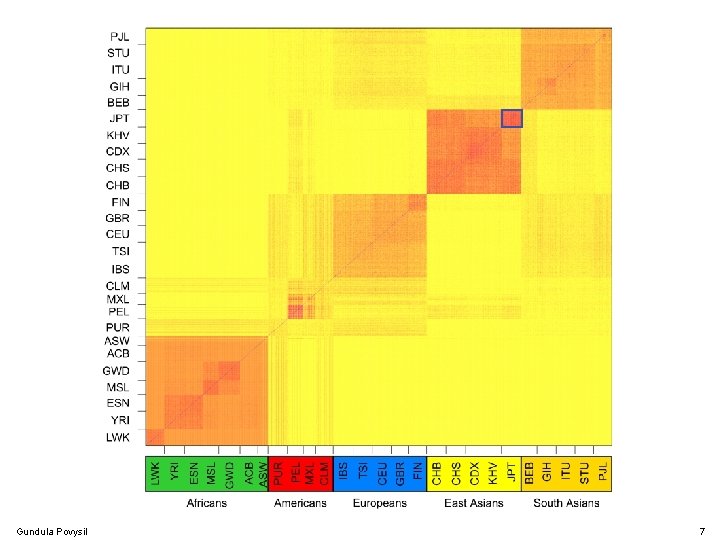
Population Structure Gundula Povysil 7
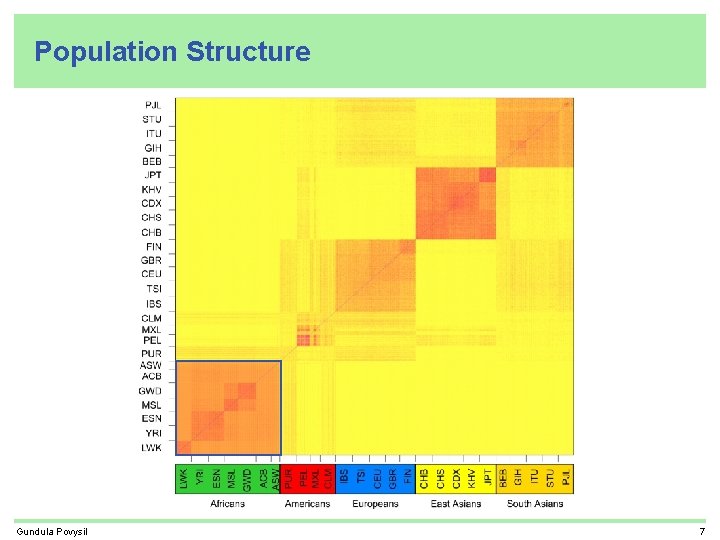
Population Structure Gundula Povysil 7
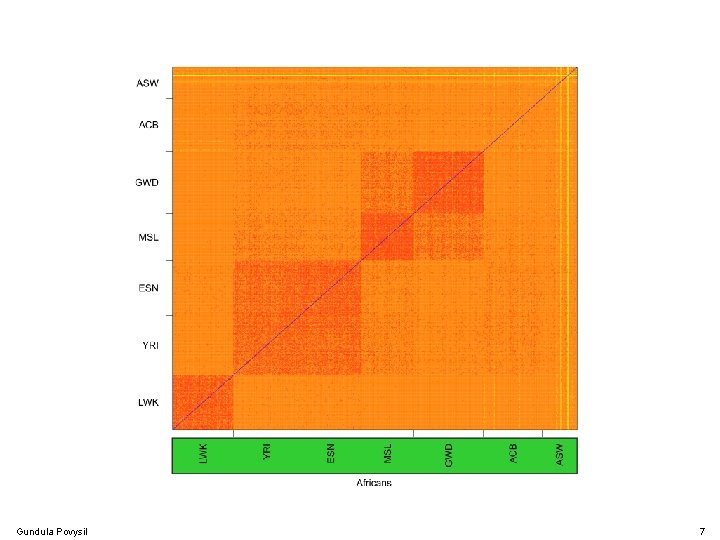
Gundula Povysil 7
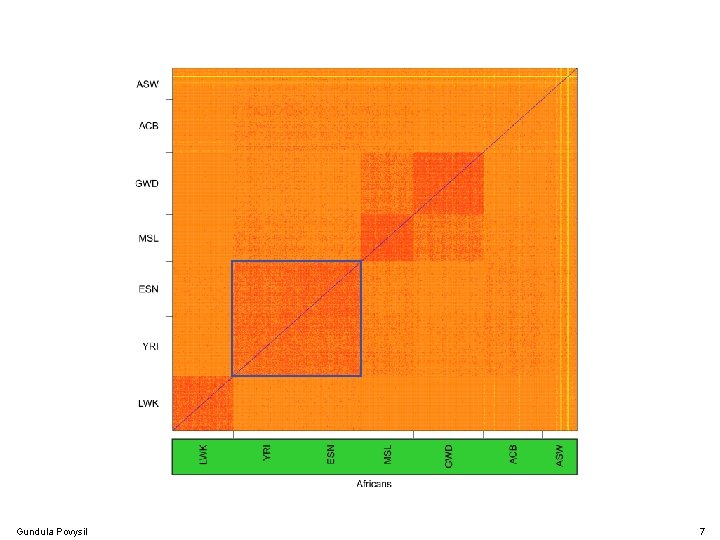
Gundula Povysil 7
Gundula Povysil 7
Gundula Povysil 7
Gundula Povysil 7
Gundula Povysil 7
Gundula Povysil 7
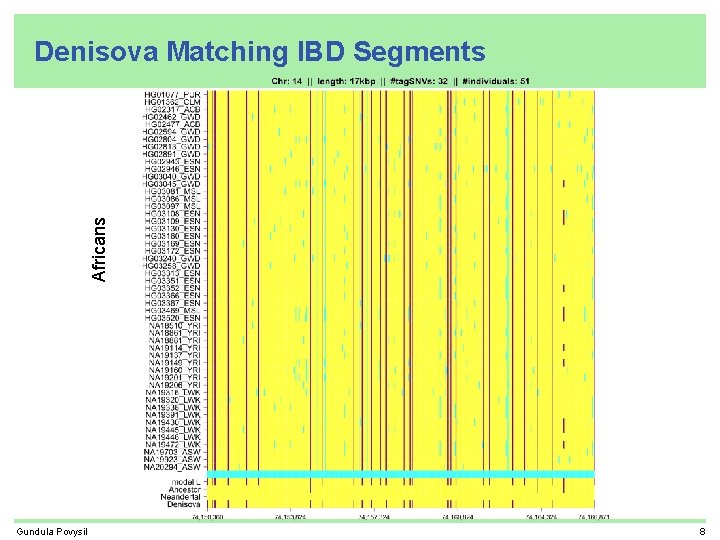
Africans Denisova Matching IBD Segments Gundula Povysil 8
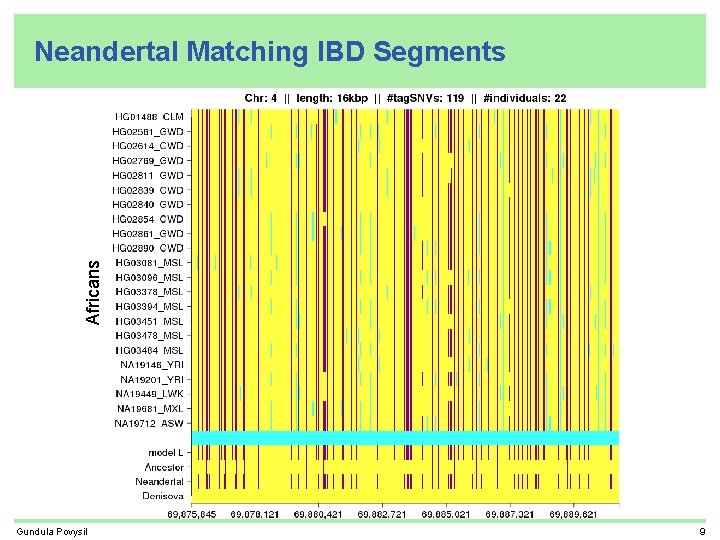
Africans Neandertal Matching IBD Segments Gundula Povysil 9
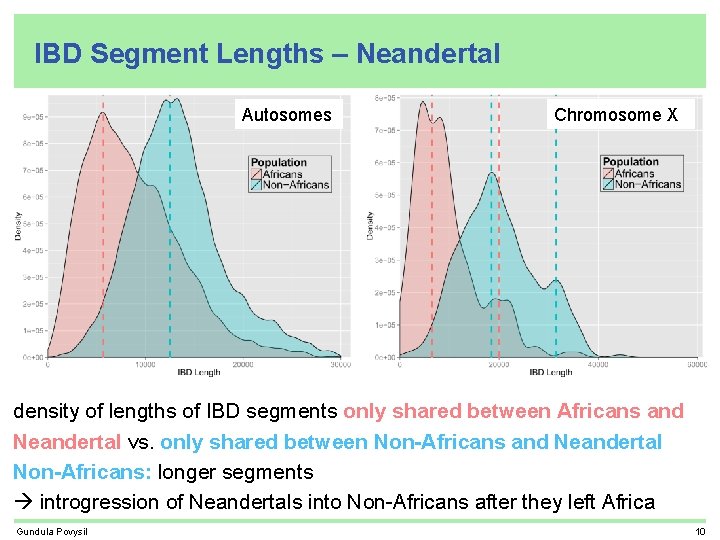
IBD Segment Lengths – Neandertal Autosomes Chromosome X density of lengths of IBD segments only shared between Africans and Neandertal vs. only shared between Non-Africans and Neandertal Non-Africans: longer segments introgression of Neandertals into Non-Africans after they left Africa Gundula Povysil 10
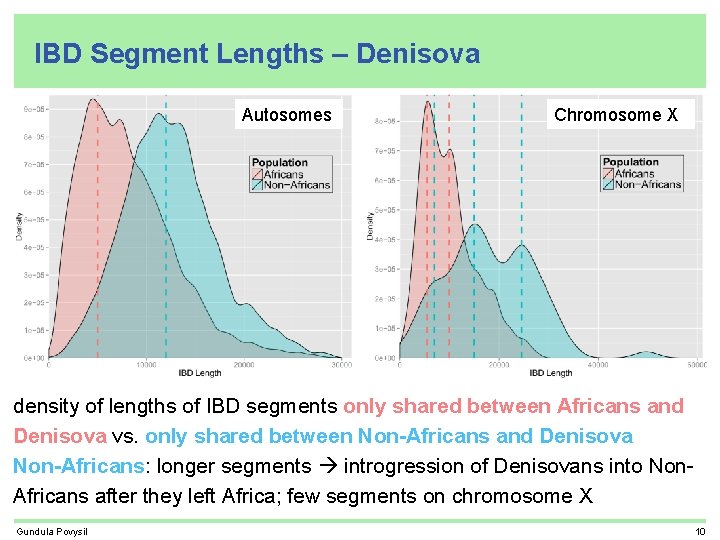
IBD Segment Lengths – Denisova Autosomes Chromosome X density of lengths of IBD segments only shared between Africans and Denisova vs. only shared between Non-Africans and Denisova Non-Africans: longer segments introgression of Denisovans into Non. Africans after they left Africa; few segments on chromosome X Gundula Povysil 10

Recap • related individuals found by IBD sharing • most IBD segments shared by Africans • many IBD segments shared by more than one super-population • each African individual carries about 10 times more IBD segments than an individual from another super-population (excluding ASW, ACB, and AMR who are all partly African) • IBD sharing helps to find population structure • IBD segments shared between Africans and Neandertal/Denisova are shorter older than IBD segments shared between Non-Africans and Neandertal/Denisova Gundula Povysil 12
Thank you! Hochreiter Group: • Sepp Hochreiter • Ulrich Bodenhofer • Günter Klambauer • Djork-Arné Clevert • Andreas Mayr povysil@bioinf. jku. at www. bioinf. jku. at • Andreas Mitterecker • Karin Schwarzbauer • Thomas Unterthiner Gundula Povysil 13
- Slides: 37