Hybridization capture highthroughput sequencing and its implications for

Hybridization capture, high-throughput sequencing and its implications for ancient DNA research Michael Hofreiter

Is science becoming infantilized? Our young people are undisciplined and sleazy. They do not listen to their parents anymore. The end of the world is near. Ur, Chaldäa, 2, 000 BC

Is ancient DNA research infantile? It’s a zebra Higuchi et al. 1984, Nature
However. . . Watson and Crick 1953 was also a short Nature paper
Simple stories are not always bad There are lies for children and lies for adults Terry Pratchett

Some more reflections Not all investigations deserve equal respect. Observations alone do not always make sense. What do we really learn from genomic data? How to win a Nobel prize? I don’t know.
From no data to drowning in data

The latest fancy piece of kit ~ 200 Gb total sequence ~ 1 billion individual reads
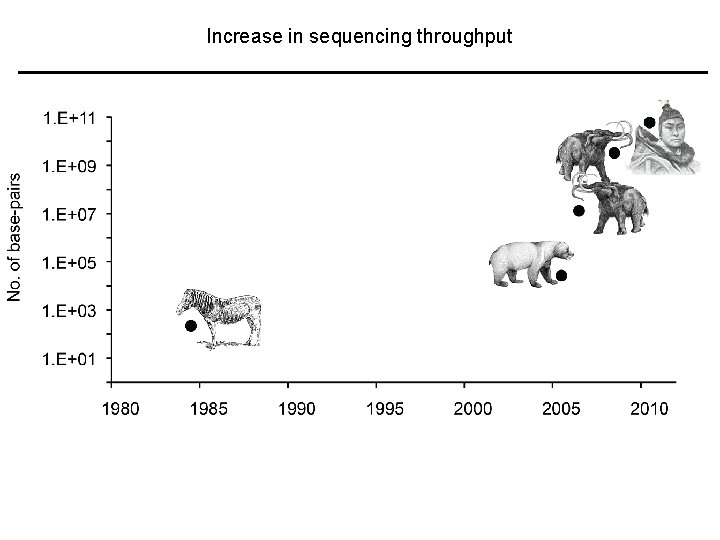
The latest throughput Increase in sequencing

Mammoth

Palaeo-Eskimos
Neanderthals

Data first?

So what have we learned from ancient genomes Mammoth genome draft: Hm. . . Saqqaq genome: Migrated from Arctic north-east Asia 5, 500 B. P. Neanderthal genome draft: Diverged from modern humans ~ 0. 4 mya Maybe gene flow into modern human gene pool Genetic regions were selected on human lineage

And what did they cost? Mammoth genome draft: ~ $ 800, 000 Saqqaq genome: $ 500, 000 Neanderthal genome draft: $ 6. 4 million

The disadvantages Shotgun sequencing Made for a maximum of 8 samples Costs - $ 20, 000 per run
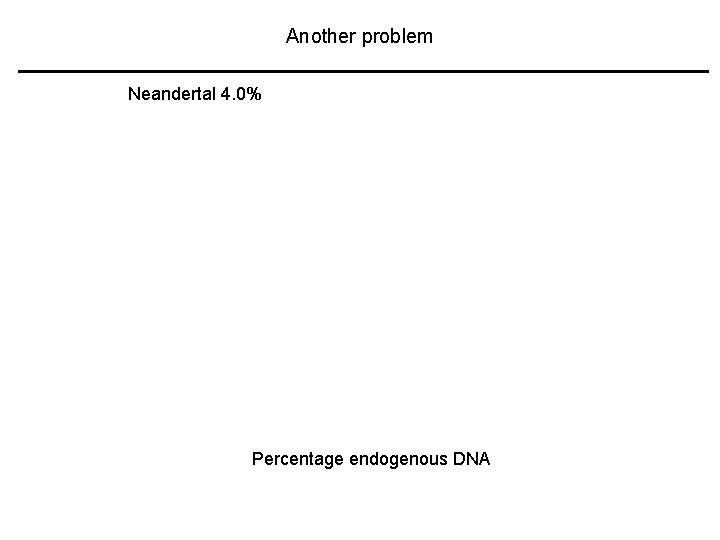
Another problem Neandertal 4. 0% Percentage endogenous DNA


Are more data better data?
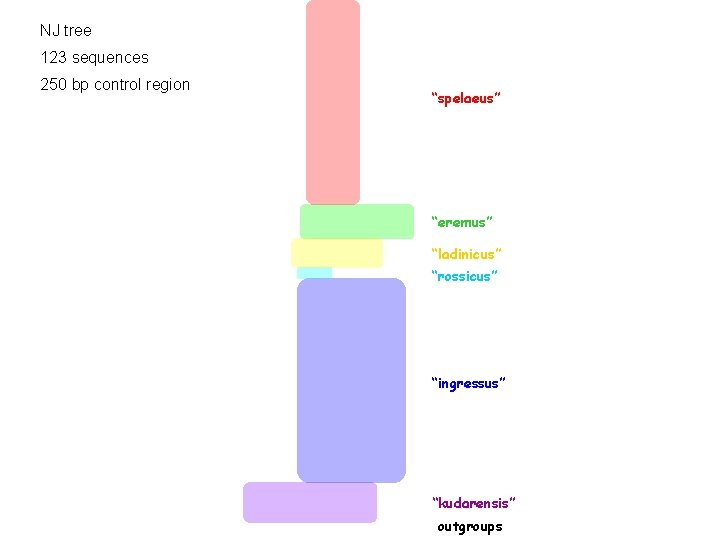
NJ tree 123 sequences 250 bp control region “spelaeus” “eremus” “ladinicus” “rossicus” “ingressus” “kudarensis” outgroups
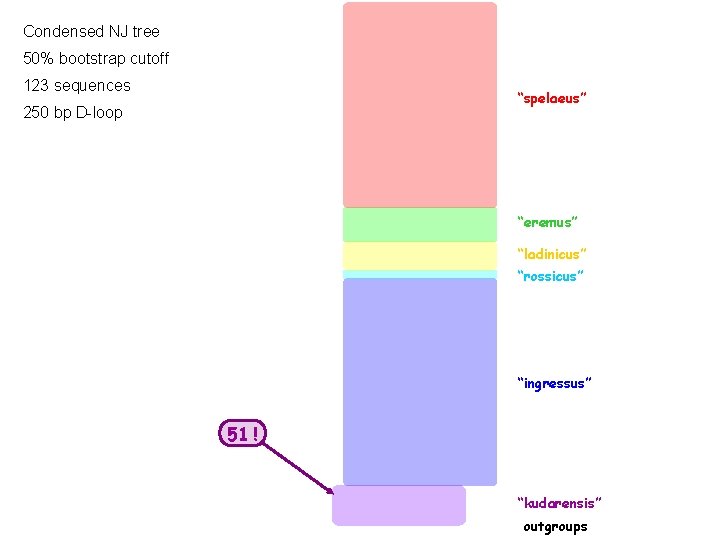
Condensed NJ tree 50% bootstrap cutoff 123 sequences “spelaeus” 250 bp D-loop “eremus” “ladinicus” “rossicus” “ingressus” 51 ! “kudarensis” outgroups
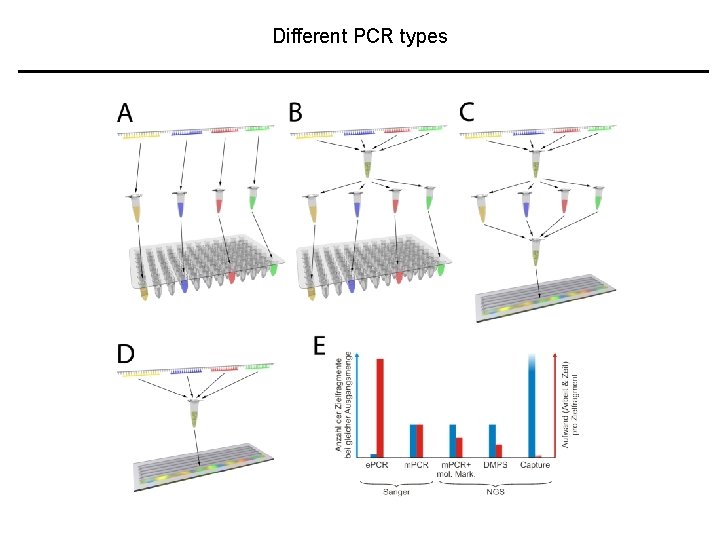
Different PCR types
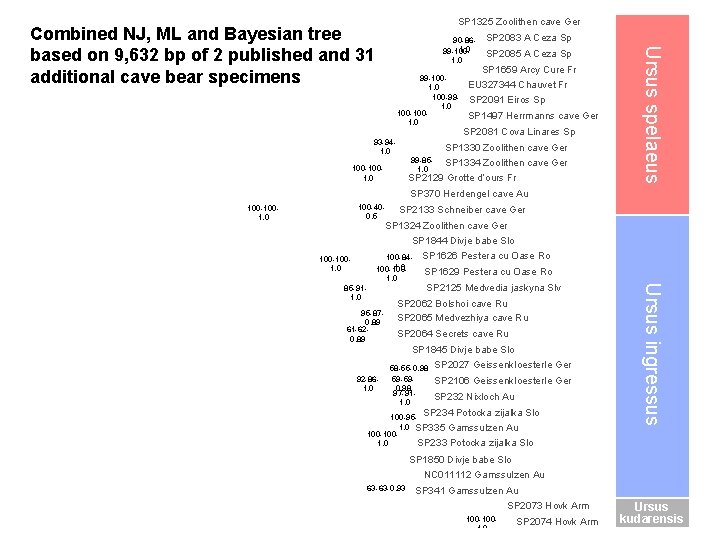
SP 1325 Zoolithen cave Ger 90 -861. 0 99 -1001. 0 100 -991. 0 100 -1001. 0 93 -941. 0 SP 2083 A Ceza Sp SP 2085 A Ceza Sp SP 1659 Arcy Cure Fr EU 327344 Chauvet Fr SP 2091 Eiros Sp SP 1497 Herrmanns cave Ger SP 2081 Cova Linares Sp SP 1330 Zoolithen cave Ger 99 -851. 0 100 -1001. 0 SP 1334 Zoolithen cave Ger SP 2129 Grotte d’ours Fr Ursus spelaeus Combined NJ, ML and Bayesian tree based on 9, 632 bp of 2 published and 31 additional cave bear specimens SP 370 Herdengel cave Au 100 -400. 5 100 -1001. 0 SP 2133 Schneiber cave Ger SP 1324 Zoolithen cave Ger SP 1844 Divje babe Slo 85 -911. 0 95 -870. 89 61 -620. 89 SP 1626 Pestera cu Oase Ro SP 1629 Pestera cu Oase Ro SP 2125 Medvedia jaskyna Slv SP 2062 Bolshoi cave Ru SP 2065 Medvezhiya cave Ru SP 2064 Secrets cave Ru SP 1845 Divje babe Slo 92 -861. 0 58 -55 -0. 98 59 -590. 98 97 -911. 0 SP 2027 Geissenkloesterle Ger SP 2106 Geissenkloesterle Ger SP 232 Nixloch Au SP 234 Potocka zijalka Slo 100 -951. 0 SP 335 Gamssulzen Au 100 -100 SP 233 Potocka zijalka Slo 1. 0 Ursus ingressus 100 -841. 0 100 -1001. 0 SP 1850 Divje babe Slo NC 011112 Gamssulzen Au 63 -63 -0. 93 SP 341 Gamssulzen Au SP 2073 Hovk Arm 100 -1001. 0 SP 2074 Hovk Arm Ursus kudarensis
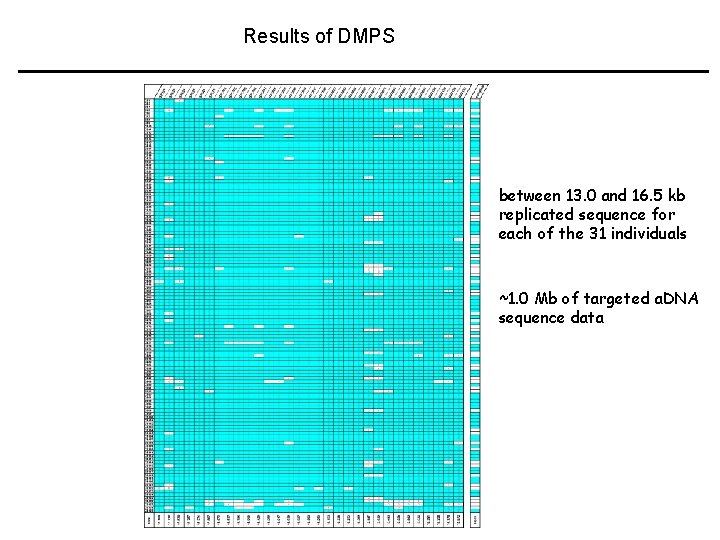
Results of DMPS between 13. 0 and 16. 5 kb replicated sequence for each of the 31 individuals ~1. 0 Mb of targeted a. DNA sequence data
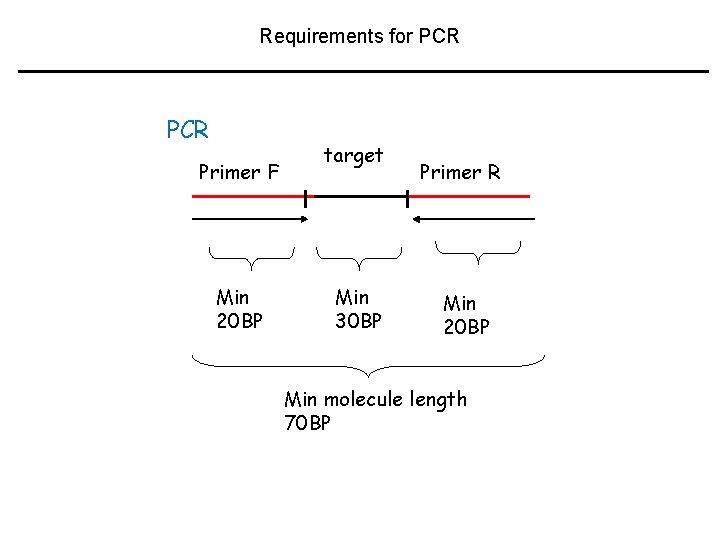
Requirements for PCR Primer F Min 20 BP target Min 30 BP Primer R Min 20 BP Min molecule length 70 BP
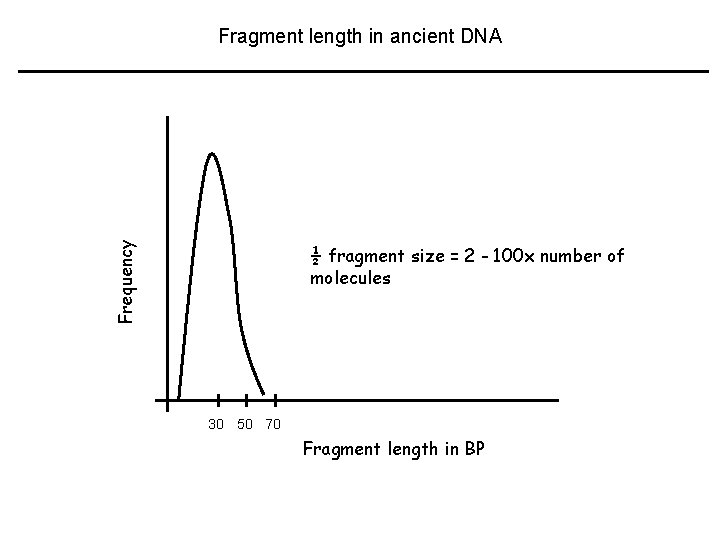
Frequency Fragment length in ancient DNA ½ fragment size = 2 - 100 x number of molecules 30 50 70 Fragment length in BP
DNA hybridization capture
DNA hybridization capture
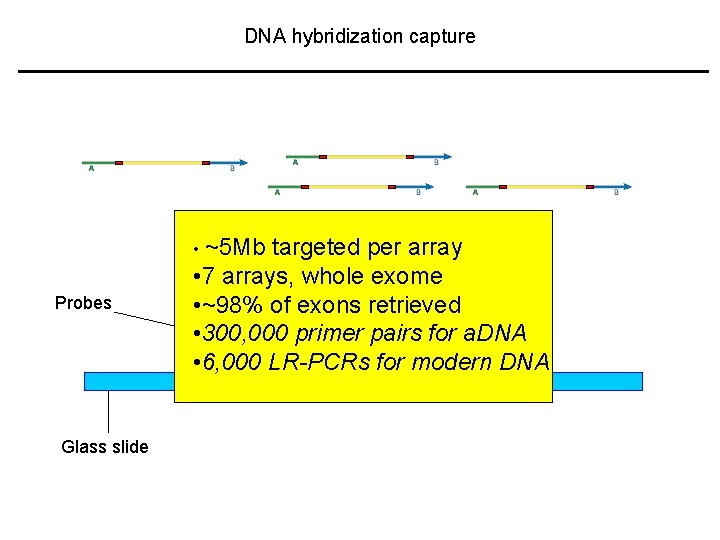
DNA hybridization capture • ~5 Mb Probes Glass slide targeted per array • 7 arrays, whole exome • ~98% of exons retrieved • 300, 000 primer pairs for a. DNA • 6, 000 LR-PCRs for modern DNA
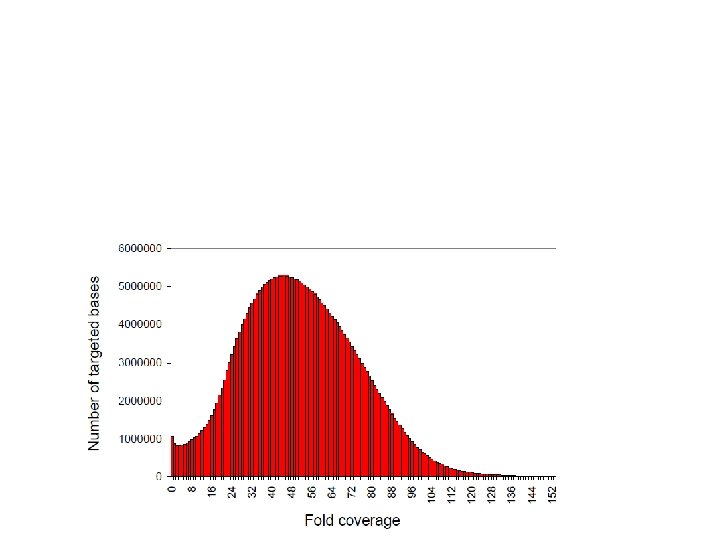
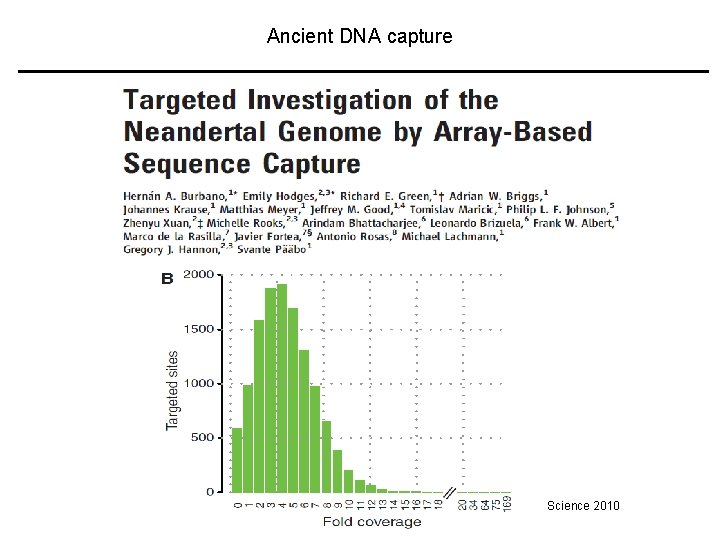
Ancient DNA capture Science 2010
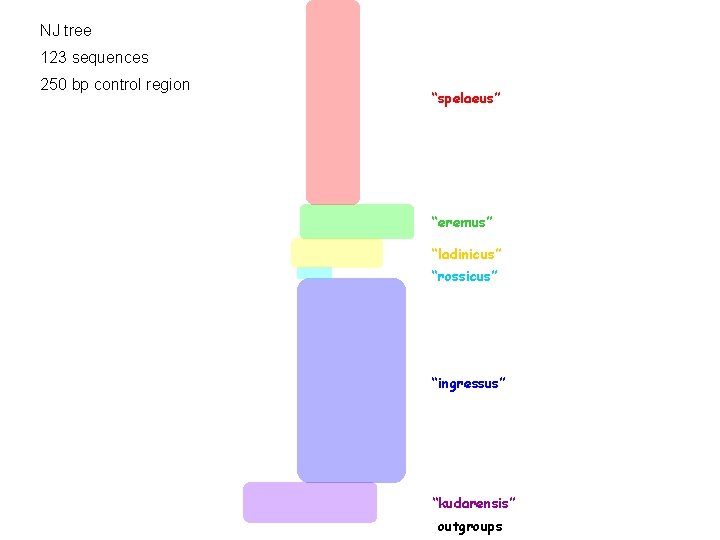
NJ tree 123 sequences 250 bp control region “spelaeus” “eremus” “ladinicus” “rossicus” “ingressus” “kudarensis” outgroups
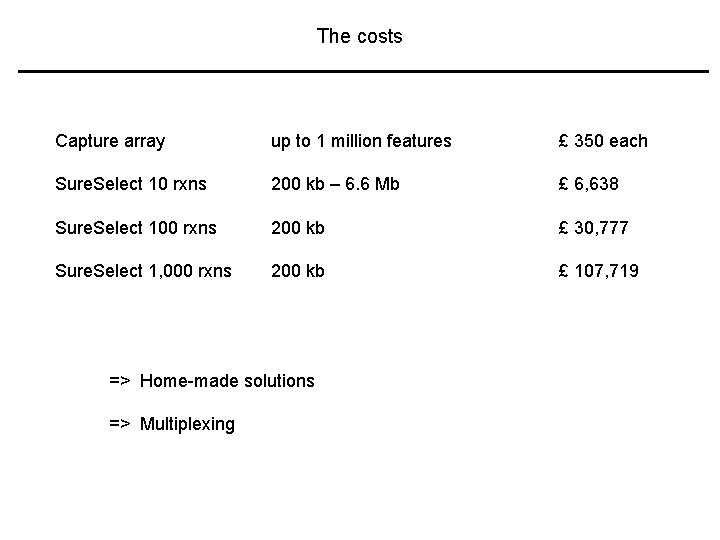
The costs Capture array up to 1 million features £ 350 each Sure. Select 10 rxns 200 kb – 6. 6 Mb £ 6, 638 Sure. Select 100 rxns 200 kb £ 30, 777 Sure. Select 1, 000 rxns 200 kb £ 107, 719 => Home-made solutions => Multiplexing
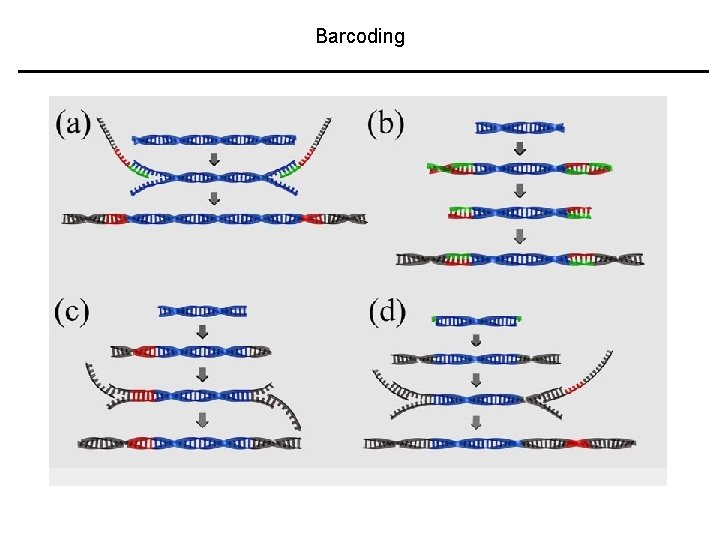
Barcoding

So. . . . . How does it work?

Sometimes well
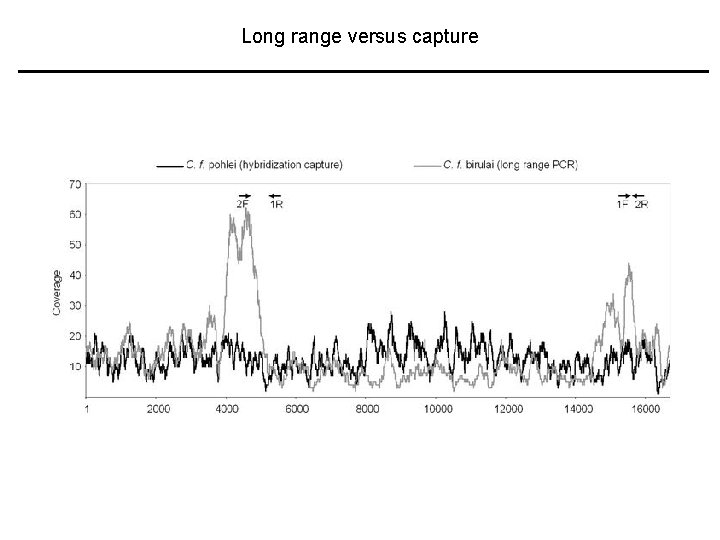
Long range versus capture

And sometimes not so well
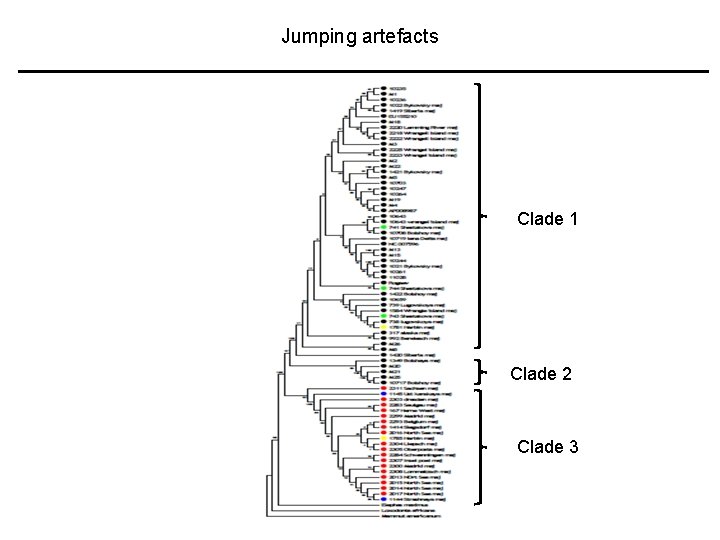
Jumping artefacts Clade 1 Clade 2 Clade 3
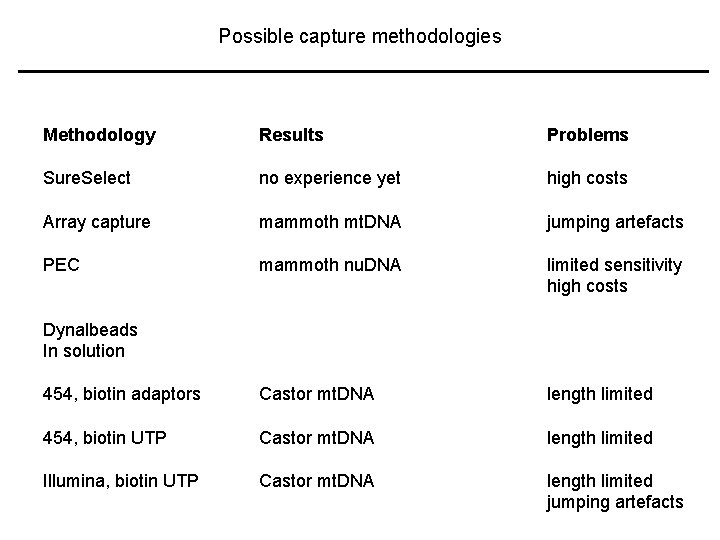
Possible capture methodologies Methodology Results Problems Sure. Select no experience yet high costs Array capture mammoth mt. DNA jumping artefacts PEC mammoth nu. DNA limited sensitivity high costs 454, biotin adaptors Castor mt. DNA length limited 454, biotin UTP Castor mt. DNA length limited Illumina, biotin UTP Castor mt. DNA length limited jumping artefacts Dynalbeads In solution
Capture advantages High sequence yield per sample aliquot Time and work efficient Higher sensitivity than PCR
Capture disadvantages High costs Sometimes low on-target ratio Problems with multiplexing Generally jumping artefacts
Summary for capture Long term little alternative - if large amounts of data required Also some methods have better sensitivity than PCR Multiplex problems especially for low-complexity data need resolving Currently not suitable for routine applications Methodological development required

Some final thoughts How should blank controls be done? And how many? What does contamination mean when you have 20 million sequence reads? How shall we replicate the data? Is independent replication possible? And is it necessary?

Thanks Molecular Ecology • Many people • Adrian Briggs, Harvard Medical school • Kevin Campbell, University of Manitoba • Research Group Molecular Ecology • Sequencing group in Leipzig • MPG, DFG and Volkswagen foundation for money • University of York • For your attention

- Slides: 46