How to Best Utilize your QEQE Plus for

How to Best Utilize your QE/QE Plus for Maximum Peptides IDs and for Peptide Quantitation Tara Schroeder Josh Nicklay Scott Peterman Yue Xuan
Part 1: Bottom Up Proteomics
Key Elements to Bottom Up Proteomics • Reliable HPLC • Sensitive column • Stable spray • Clean samples • Optimized instrument method • Fast and accurate data processing software We will focus here! 3
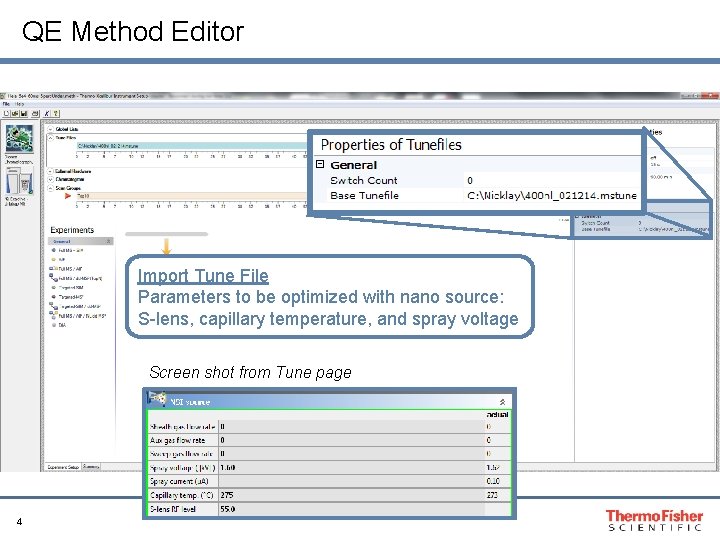
QE Method Editor Import Tune File Parameters to be optimized with nano source: S-lens, capillary temperature, and spray voltage Screen shot from Tune page 4
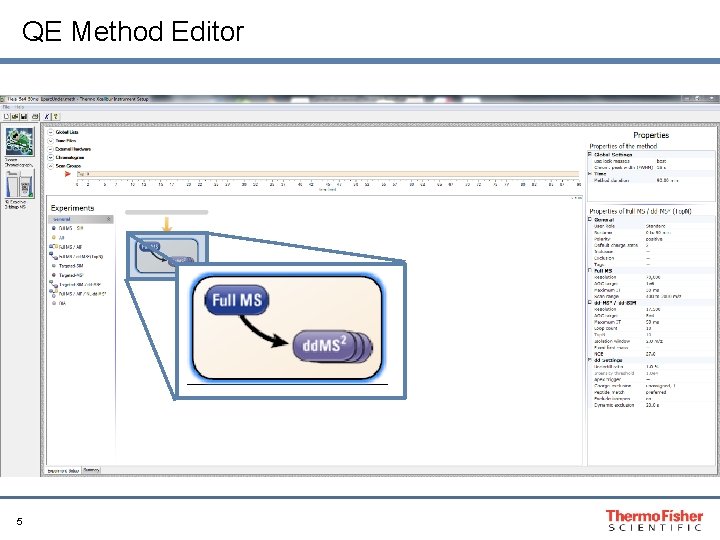
QE Method Editor 5
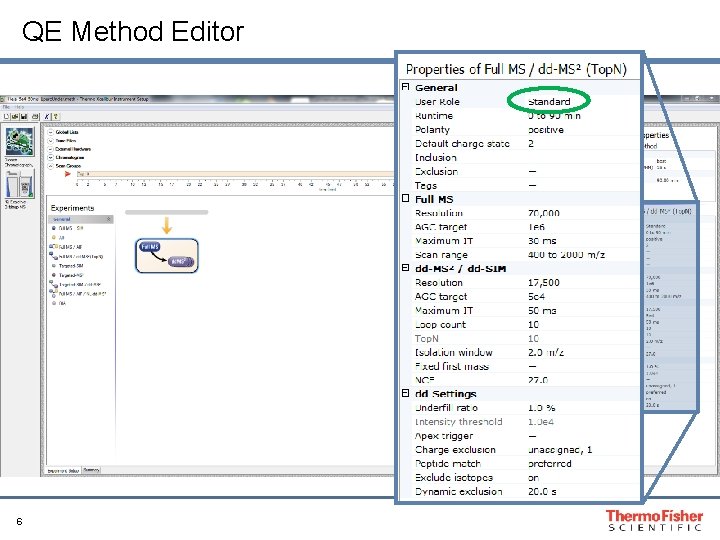
QE Method Editor 6
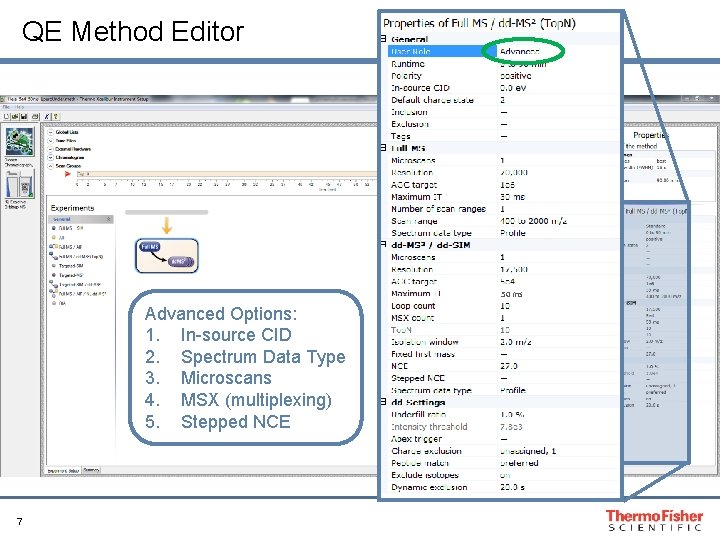
QE Method Editor 50 ms Advanced Options: 1. In-source CID 2. Spectrum Data Type 3. Microscans 4. MSX (multiplexing) 5. Stepped NCE 7
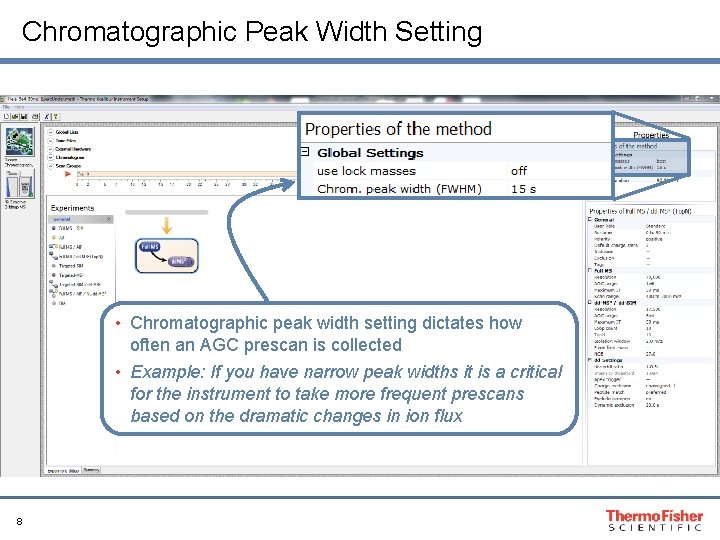
Chromatographic Peak Width Setting • Chromatographic peak width setting dictates how often an AGC prescan is collected • Example: If you have narrow peak widths it is a critical for the instrument to take more frequent prescans based on the dramatic changes in ion flux 8
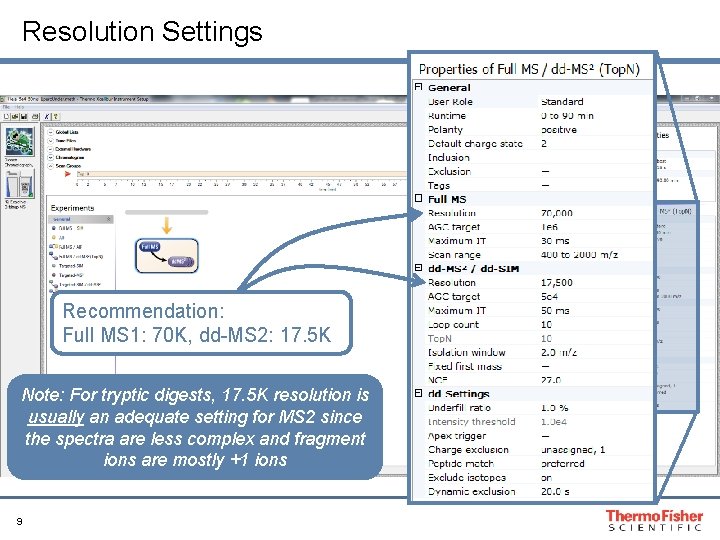
Resolution Settings Recommendation: Full MS 1: 70 K, dd-MS 2: 17. 5 K Note: For tryptic digests, 17. 5 K resolution is usually an adequate setting for MS 2 since the spectra are less complex and fragment ions are mostly +1 ions 9
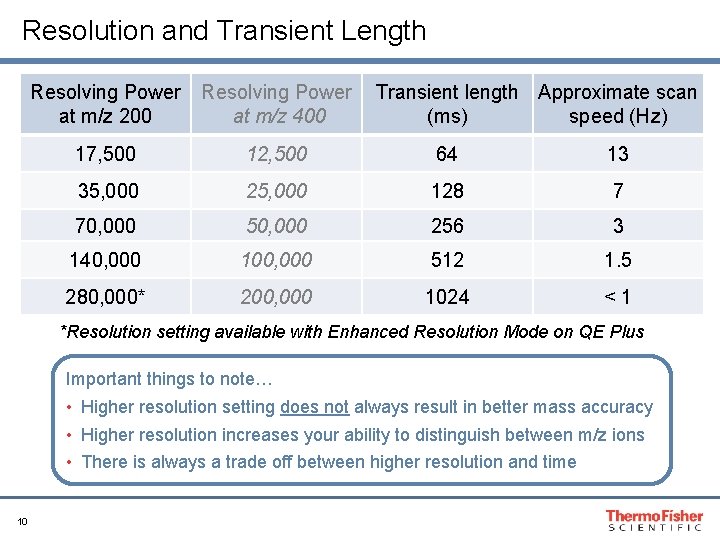
Resolution and Transient Length Resolving Power at m/z 200 Resolving Power at m/z 400 Transient length Approximate scan (ms) speed (Hz) 17, 500 12, 500 64 13 35, 000 25, 000 128 7 70, 000 50, 000 256 3 140, 000 100, 000 512 1. 5 280, 000* 200, 000 1024 <1 *Resolution setting available with Enhanced Resolution Mode on QE Plus Important things to note… • Higher resolution setting does not always result in better mass accuracy • Higher resolution increases your ability to distinguish between m/z ions • There is always a trade off between higher resolution and time 10
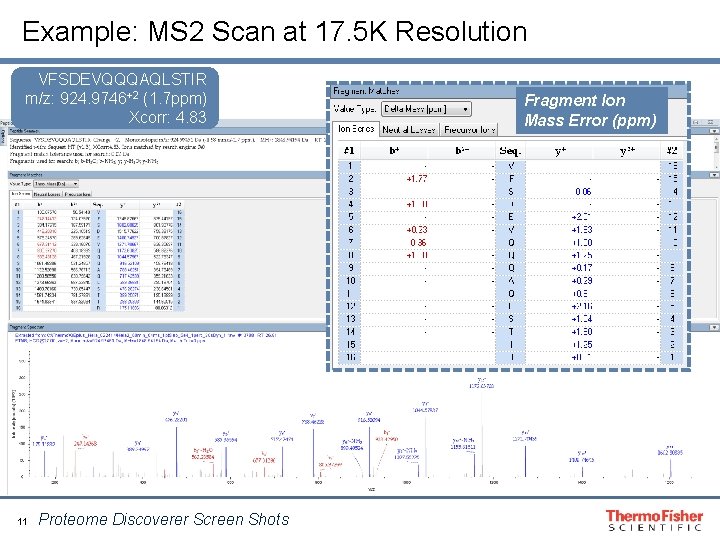
Example: MS 2 Scan at 17. 5 K Resolution VFSDEVQQQAQLSTIR m/z: 924. 9746+2 (1. 7 ppm) Xcorr: 4. 83 11 Proteome Discoverer Screen Shots Fragment Ion Mass Error (ppm)
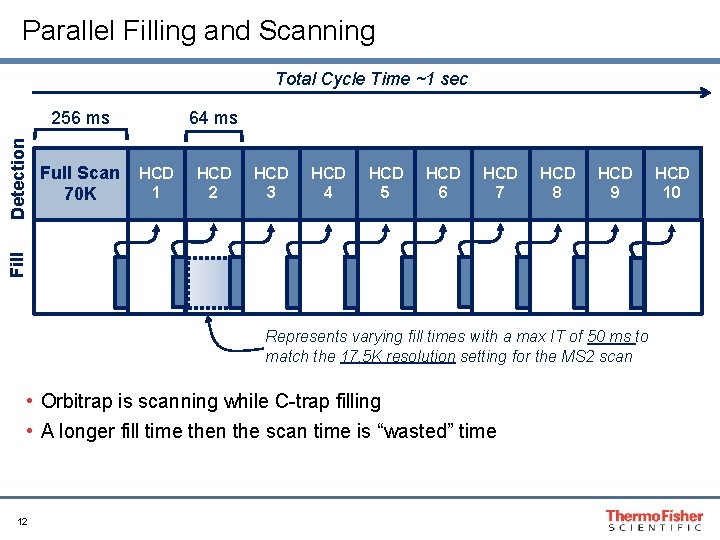
Parallel Filling and Scanning Total Cycle Time ~1 sec Full Scan 70 K 64 ms HCD 1 HCD 2 HCD 3 HCD 4 HCD 5 HCD 6 HCD 7 HCD 8 HCD 9 Fill Detection 256 ms Represents varying fill times with a max IT of 50 ms to match the 17. 5 K resolution setting for the MS 2 scan • Orbitrap is scanning while C-trap filling • A longer fill time then the scan time is “wasted” time 12 HCD 10

Suggested Max Fill Time Resolving Power Approximate scan Transient length Suggested Max Fill at m/z 200 speed (Hz) (ms) Time (ms) 17, 500 13 64 50 35, 000 7 128 110 70, 000 3 256 240 140, 000 1. 5 512 500 • Balance the max fill time with the transient scan time for the resolution to make the most effective use of the parallel fill and detect capabilities of the QE 13
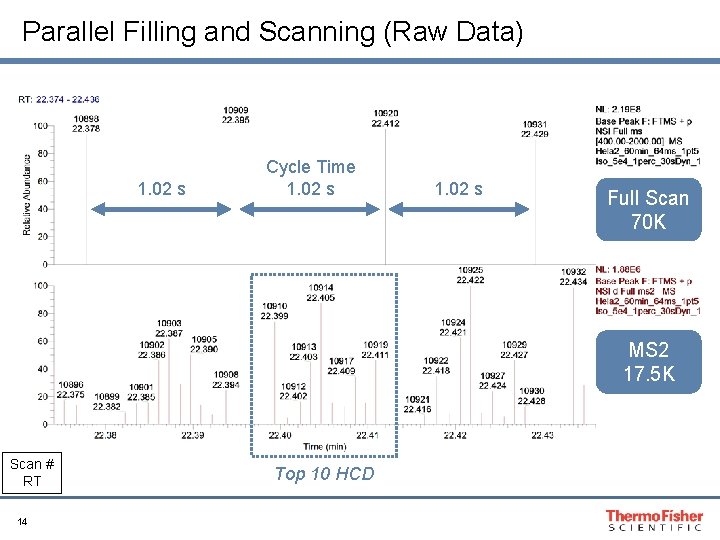
Parallel Filling and Scanning (Raw Data) 1. 02 s Cycle Time 1. 02 s Full Scan 70 K MS 2 17. 5 K Scan # RT 14 Top 10 HCD
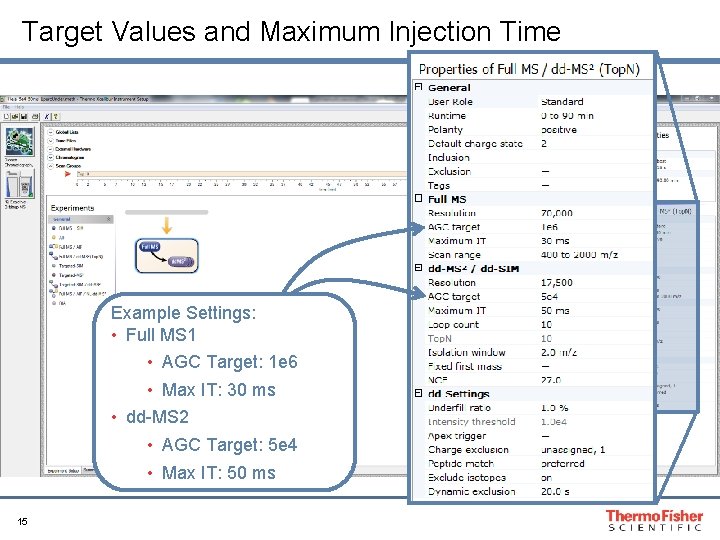
Target Values and Maximum Injection Time Example Settings: • Full MS 1 • AGC Target: 1 e 6 • Max IT: 30 ms • dd-MS 2 • AGC Target: 5 e 4 • Max IT: 50 ms 15
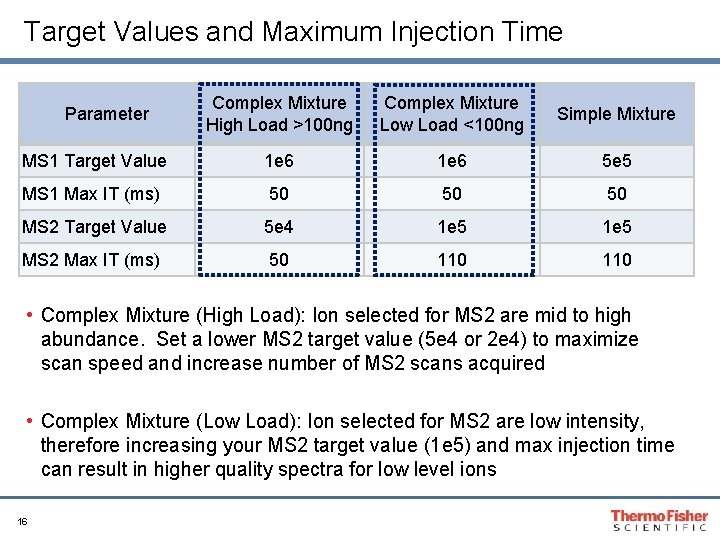
Target Values and Maximum Injection Time Complex Mixture High Load >100 ng Complex Mixture Low Load <100 ng Simple Mixture MS 1 Target Value 1 e 6 5 e 5 MS 1 Max IT (ms) 50 50 50 MS 2 Target Value 5 e 4 1 e 5 MS 2 Max IT (ms) 50 110 Parameter • Complex Mixture (High Load): Ion selected for MS 2 are mid to high abundance. Set a lower MS 2 target value (5 e 4 or 2 e 4) to maximize scan speed and increase number of MS 2 scans acquired • Complex Mixture (Low Load): Ion selected for MS 2 are low intensity, therefore increasing your MS 2 target value (1 e 5) and max injection time can result in higher quality spectra for low level ions 16
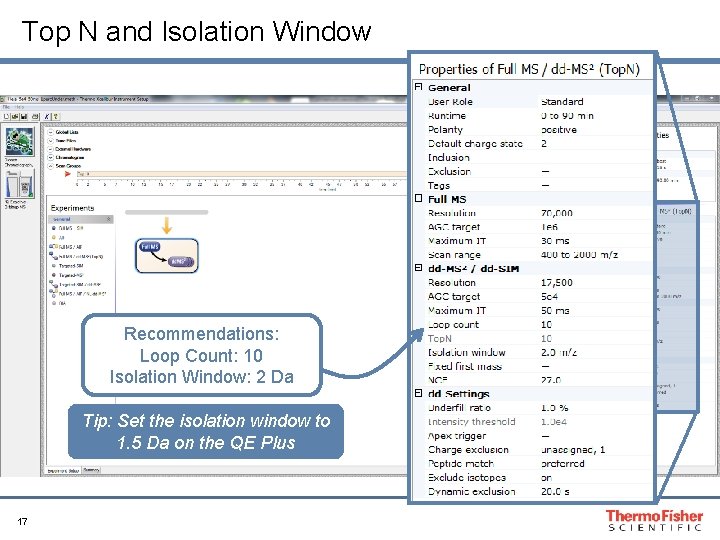
Top N and Isolation Window Recommendations: Loop Count: 10 Isolation Window: 2 Da Tip: Set the isolation window to 1. 5 Da on the QE Plus 17
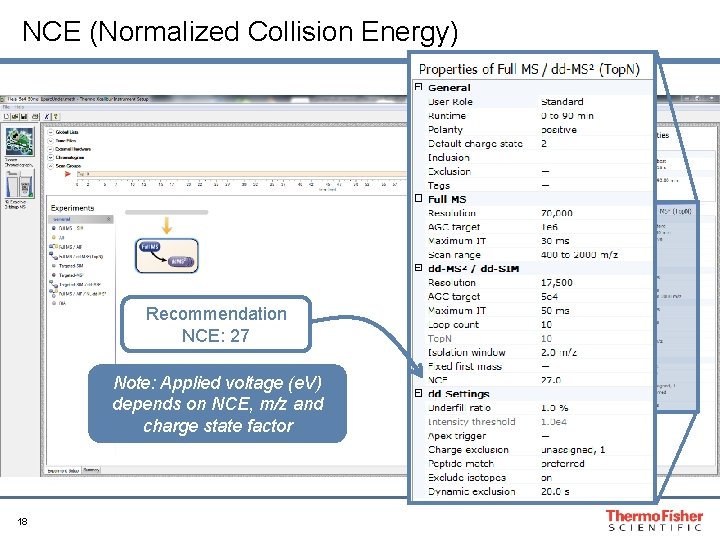
NCE (Normalized Collision Energy) Recommendation NCE: 27 Note: Applied voltage (e. V) depends on NCE, m/z and charge state factor 18

Peptide Match Recommendation: Peptide Match Preferred 19
Peptide Match • Definition: • Enables the QE to select ions with peptide-like isotopic distributions for Data Dependent scanning. • It recognizes the monoisotopic mass of an isotopic distribution by comparing isotopic intensity ratios to typical peptide-like distributions. • 3 Options: • On: will only select ions for dd-MS 2 if recognized as having a peptide-like isotopic distribution • “–”: isotopic pattern will have no affect on MS 2 selection • Preferred: ions with peptide isotopic pattern are triggered with preference to ions with peptide match. However, after it selects all ions that have “peptide match” it will continue to select other ions for dd-MS 2 Tip: When analyzing a complex mixture, setting peptide match to preferred will maximize the number of MS 2 scans collected 20 Tip: When analyzing a simple mixture, consider turning Peptide Match “off”
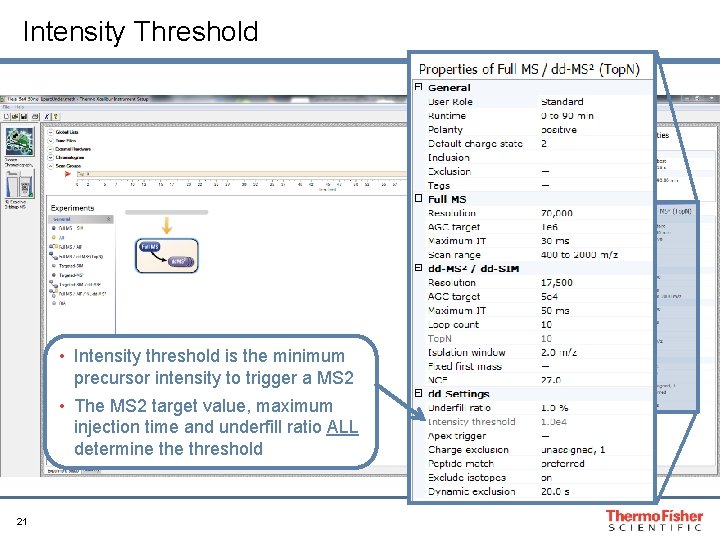
Intensity Threshold • Intensity threshold is the minimum precursor intensity to trigger a MS 2 • The MS 2 target value, maximum injection time and underfill ratio ALL determine threshold 21
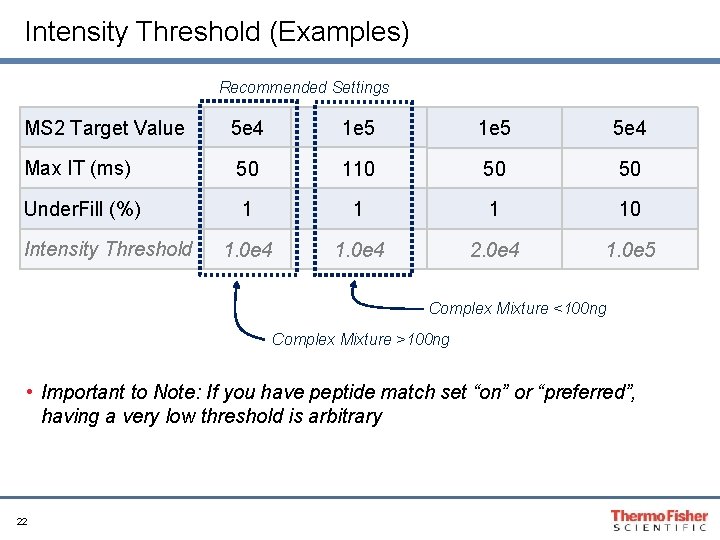
Intensity Threshold (Examples) Recommended Settings MS 2 Target Value 5 e 4 1 e 5 5 e 4 Max IT (ms) 50 110 50 50 Under. Fill (%) 1 10 1. 0 e 4 2. 0 e 4 1. 0 e 5 Intensity Threshold Complex Mixture <100 ng Complex Mixture >100 ng • Important to Note: If you have peptide match set “on” or “preferred”, having a very low threshold is arbitrary 22
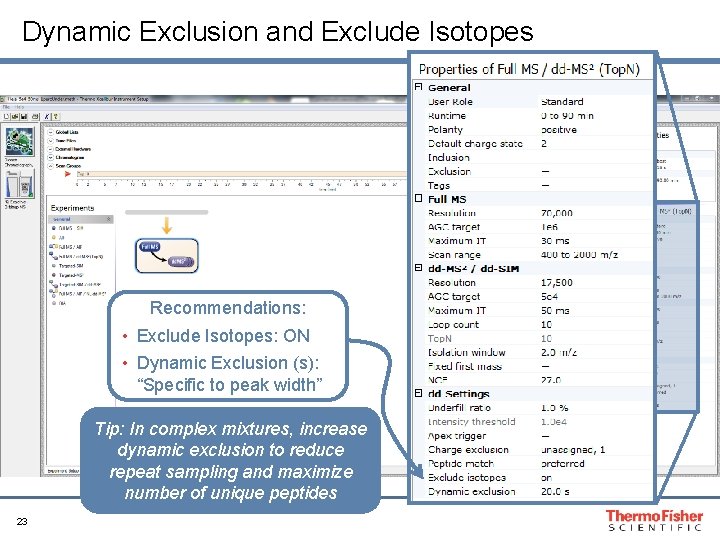
Dynamic Exclusion and Exclude Isotopes Recommendations: • Exclude Isotopes: ON • Dynamic Exclusion (s): “Specific to peak width” Tip: In complex mixtures, increase dynamic exclusion to reduce repeat sampling and maximize number of unique peptides 23
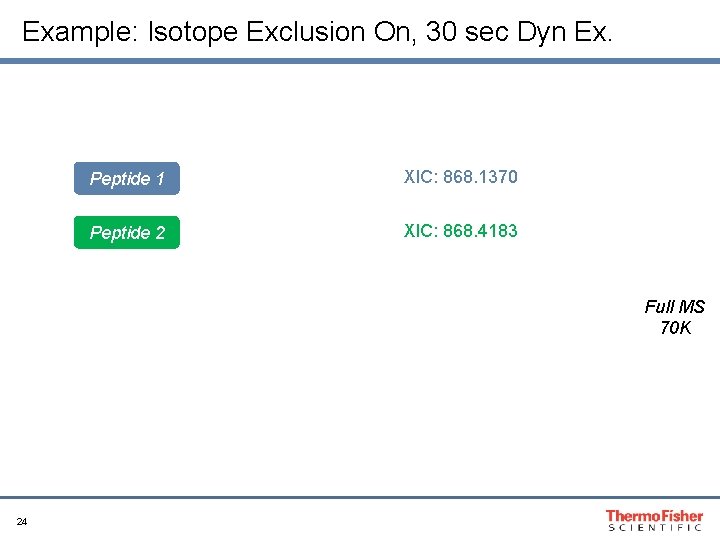
Example: Isotope Exclusion On, 30 sec Dyn Ex. Peptide 1 XIC: 868. 1370 Peptide 2 XIC: 868. 4183 Full MS 70 K 24
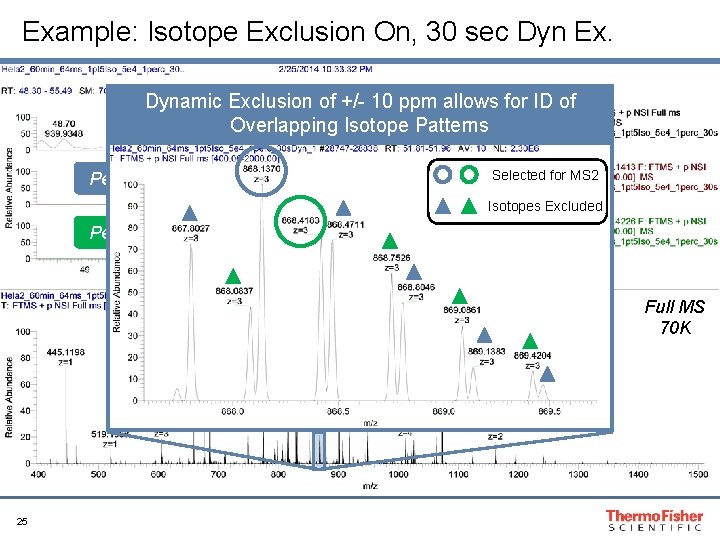
Example: Isotope Exclusion On, 30 sec Dyn Ex. Dynamic Exclusion of +/- 10 ppm allows for ID of Overlapping Isotope Patterns Peptide 1 Selected for MS 2 Isotopes Excluded Peptide 2 Full MS 70 K 25
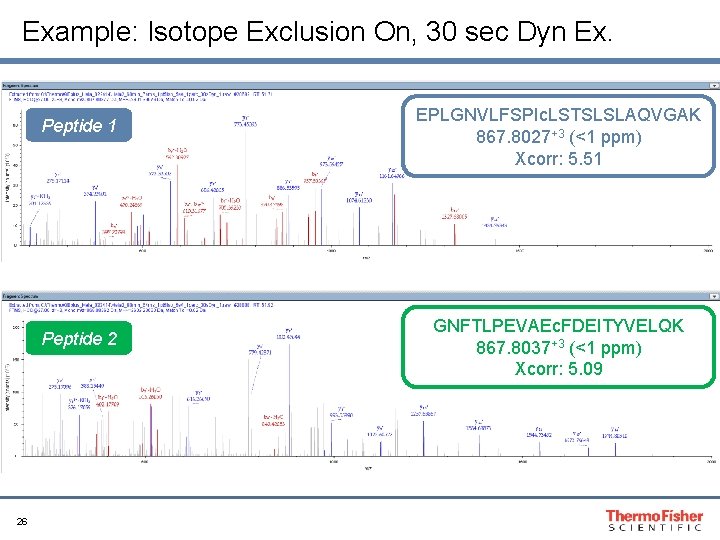
Example: Isotope Exclusion On, 30 sec Dyn Ex. Peptide 1 Peptide 2 26 EPLGNVLFSPIc. LSTSLSLAQVGAK 867. 8027+3 (<1 ppm) Xcorr: 5. 51 GNFTLPEVAEc. FDEITYVELQK 867. 8037+3 (<1 ppm) Xcorr: 5. 09
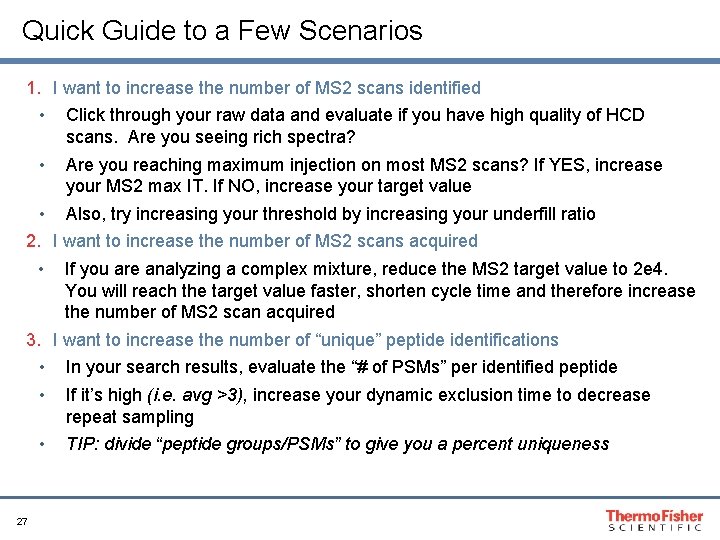
Quick Guide to a Few Scenarios 1. I want to increase the number of MS 2 scans identified • Click through your raw data and evaluate if you have high quality of HCD scans. Are you seeing rich spectra? • Are you reaching maximum injection on most MS 2 scans? If YES, increase your MS 2 max IT. If NO, increase your target value • Also, try increasing your threshold by increasing your underfill ratio 2. I want to increase the number of MS 2 scans acquired • If you are analyzing a complex mixture, reduce the MS 2 target value to 2 e 4. You will reach the target value faster, shorten cycle time and therefore increase the number of MS 2 scan acquired 3. I want to increase the number of “unique” peptide identifications • In your search results, evaluate the “# of PSMs” per identified peptide • If it’s high (i. e. avg >3), increase your dynamic exclusion time to decrease repeat sampling • TIP: divide “peptide groups/PSMs” to give you a percent uniqueness 27
Part 2: Peptide Quantitation

Define the Experiment • What are your goals? • Are you trying to quantitate 1000 peptides? • Is absolute quantitation of 5 targets your priority? • There will always be a sacrifice within the targeted experiment • If you want extreme breadth, then you will sacrifice depth • If you want extreme depth, then you will need to decrease the number of your targets 29

Keys to a Successful Quantitation Experiment • Reproducibility!! • Reproducibility in retention time • Reproducibility in overall signal within replicates • Obtaining adequate scans across the peak • This is critical for accurate quantitation • Tip: 10 scans across the peak for a middle level target will result in enough scans across the peak at your limit of detection • Sample complexity • Interferences will dictate your limit of detection 30
Questions Before Starting Analysis • How many targets do you have? • How wide are your chromatographic peaks? • What is the complexity of your sample? The answers to these simple questions will greatly impact the design of your targeted instrument method 31
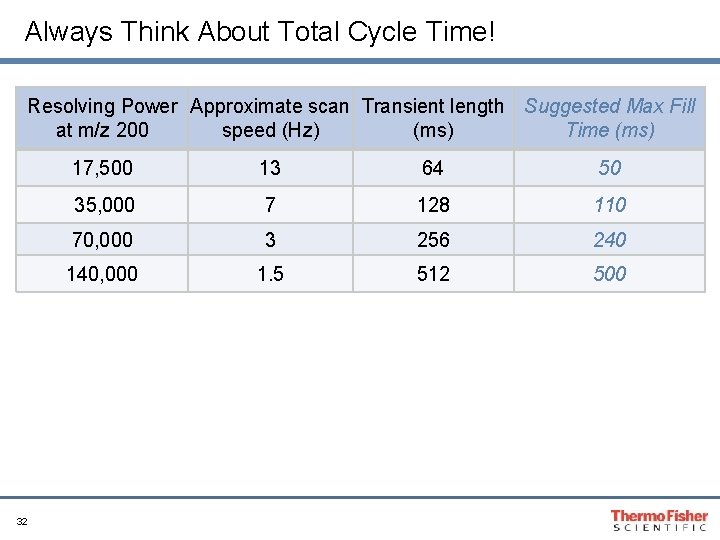
Always Think About Total Cycle Time! Resolving Power Approximate scan Transient length Suggested Max Fill at m/z 200 speed (Hz) (ms) Time (ms) 32 17, 500 13 64 50 35, 000 7 128 110 70, 000 3 256 240 140, 000 1. 5 512 500
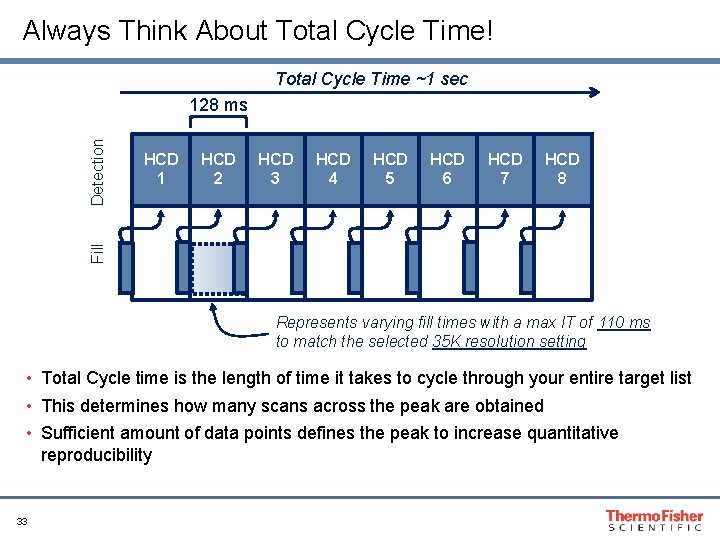
Always Think About Total Cycle Time! Total Cycle Time ~1 sec HCD 1 HCD 2 HCD 3 HCD 4 HCD 5 HCD 6 HCD 7 HCD 8 Fill Detection 128 ms Represents varying fill times with a max IT of 110 ms to match the selected 35 K resolution setting • Total Cycle time is the length of time it takes to cycle through your entire target list • This determines how many scans across the peak are obtained • Sufficient amount of data points defines the peak to increase quantitative reproducibility 33
Timed or Untimed? • How many targets do you have? • 10 or less: Can be run untimed • 10 or more: Most likely needs to be timed • How wide are your chromatographic peaks? • Example: 20 sec wide peaks • Want 10 scans across peak. Total cycle time should not exceed 2 sec. • For t. MS 2 method, using 35 K (~8 Hz), target at most 16 ions at a given time. • For SIM method, using 140 K (~2 Hz), target at most 4 ions at a given time • Example: 10 sec wide peaks • Want 10 scans across peak. Total cycle time should not exceed 1 sec. • For t. MS 2 method, using 35 K (~8 Hz), target at most 8 ions at a given time • For SIM method, using 140 K (~2 Hz), target at most 2 ions at a given time 34
Different Scan Mode Options • Quantitation by… • Full MS • SIM (Single Ion Monitoring) • t. MS 2 (Targeted MS 2) • All Scan modes utilize high resolution/accurate mass (HR/AM) • Extract ions with narrow mass window (<5 ppm) • 2 Major questions that will dictate what is the best scan mode for your experiment… • Is your priority depth or breadth? • How complex is your sample? 35
Global Quantitation: Full MS • Priority: Breadth (Very high number of targets) • Complexity: High charge density (AGC 1 e 6 to 3 e 6) • High resolution across dynamic range (140 K) • Option of acquiring confirmatory MS 2 • Qualitative Attributes: RT, accurate mass, and isotopic distribution • Quantitative Attribute: Peak areas of precursor isotopes 36
Absolute Quantitation: SIM or t. MS 2? • Priority: Sensitivity! • What is the complexity of your sample? • Simple mixture: SIM will most likely result in highest sensitivity • Peptide stays intact, achieve intense signal • Complex mixture: t. MS 2 is usually the best choice. • When targeting low level ions, SIM experiments in a complex mixture is likely limited by interferences from the matrix • Problem: Co-eluting ion in your isolation window • Symptom: Trap will faster with the interference relative to your target • Result: Decreased sensitivity of your target • t. MS 2 increases selectivity and overall improves sensitivity in the presence of co-eluting ions - Quantitation of fragment ions is more selective! 37
Scan Mode: SIM • Use the highest resolution setting to resolve your target from co-eluting species in the isolation window • Select number of isotopes for quantitation post acquisition • Option of multiplexing (MSX) to maximize cycle time • Qualitative Attributes: RT, accurate mass, and isotopic distribution • Quantitative Attribute: Peak areas of precursor isotopes 38
SIM Method 39
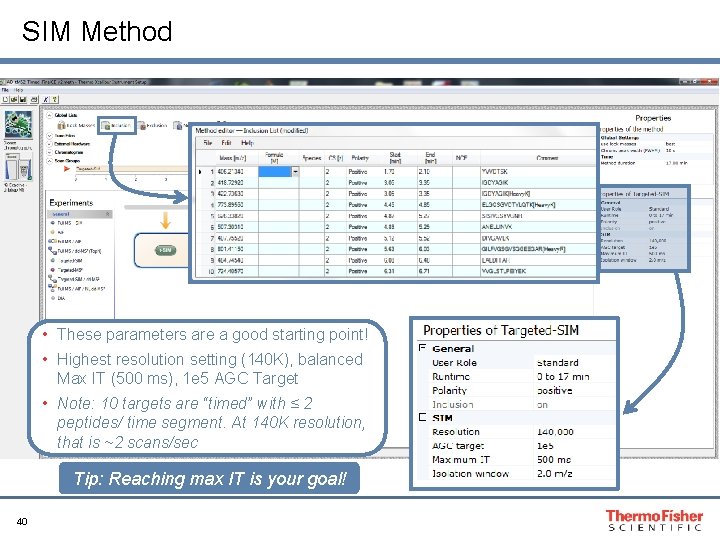
SIM Method • These parameters are a good starting point! • Highest resolution setting (140 K), balanced Max IT (500 ms), 1 e 5 AGC Target • Note: 10 targets are “timed” with ≤ 2 peptides/ time segment. At 140 K resolution, that is ~2 scans/sec Tip: Reaching max IT is your goal! 40
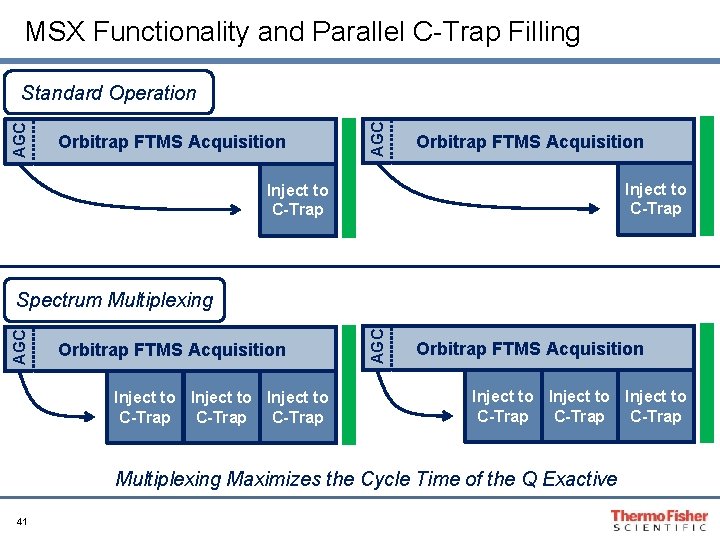
MSX Functionality and Parallel C-Trap Filling Orbitrap FTMS Acquisition AGC Standard Operation Orbitrap FTMS Acquisition Inject to C-Trap Orbitrap FTMS Acquisition Inject to C-Trap AGC Spectrum Multiplexing Orbitrap FTMS Acquisition Inject to C-Trap Multiplexing Maximizes the Cycle Time of the Q Exactive 41
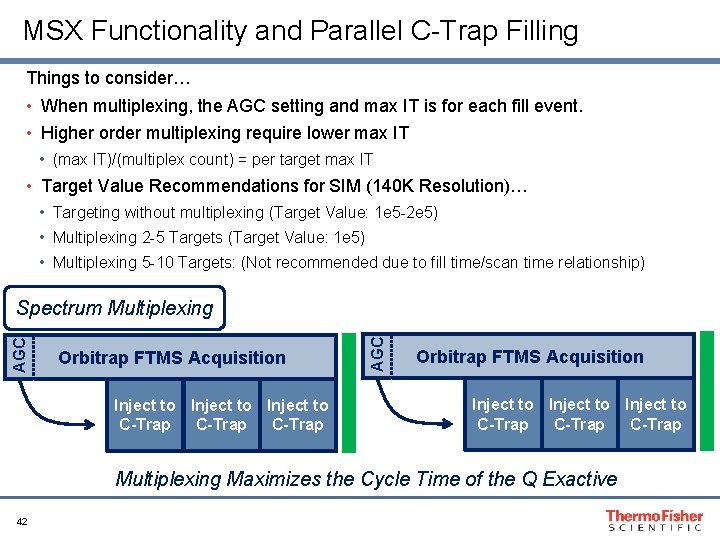
MSX Functionality and Parallel C-Trap Filling Things to consider… • When multiplexing, the AGC setting and max IT is for each fill event. • Higher order multiplexing require lower max IT • (max IT)/(multiplex count) = per target max IT • Target Value Recommendations for SIM (140 K Resolution)… • Targeting without multiplexing (Target Value: 1 e 5 -2 e 5) • Multiplexing 2 -5 Targets (Target Value: 1 e 5) • Multiplexing 5 -10 Targets: (Not recommended due to fill time/scan time relationship) Orbitrap FTMS Acquisition Inject to C-Trap AGC Spectrum Multiplexing Orbitrap FTMS Acquisition Inject to C-Trap Multiplexing Maximizes the Cycle Time of the Q Exactive 42
t. MS 2 • Generates high resolution full mass range MS 2 spectra • Parallel reaction monitoring (PRM) • Flexibility to choose the specific fragment ions for quantitation post acquisition • Since it is a high resolution scan, you can extract your ion with a narrow ppm mass window achieving high selectivity • Qualitative Attribute: RT, fragment accurate mass, and fragment ion ratios • Quantitative Attribute: Peak areas of selected fragment ions 43
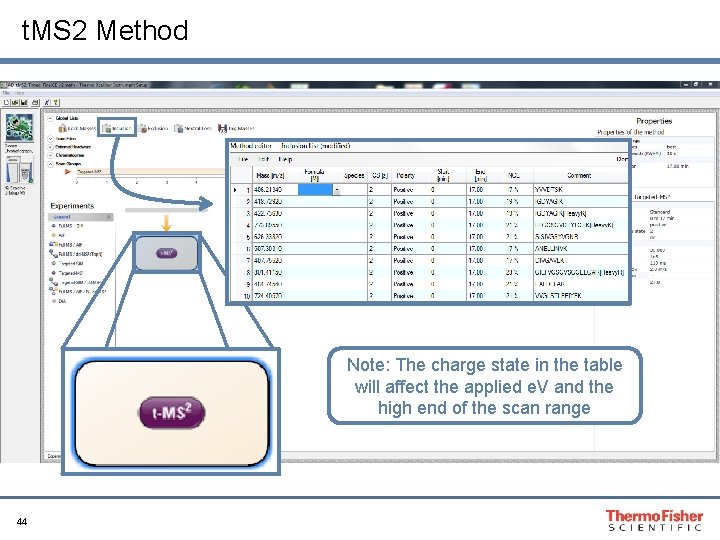
t. MS 2 Method Note: The charge state in the table will affect the applied e. V and the high end of the scan range 44
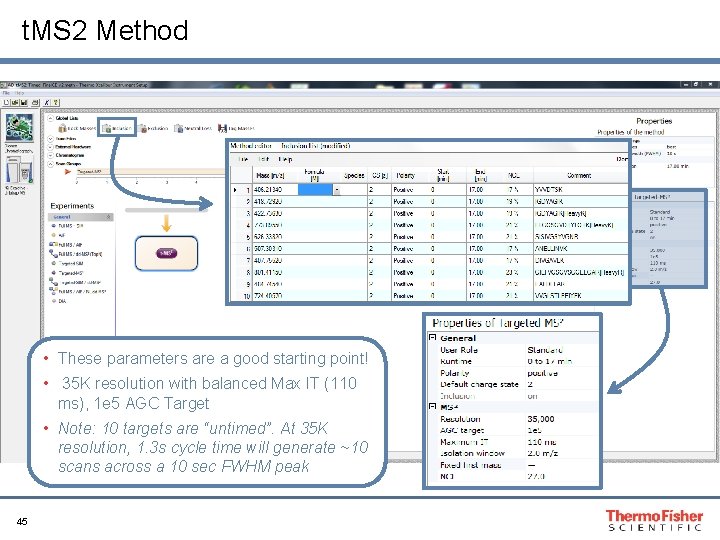
t. MS 2 Method • These parameters are a good starting point! • 35 K resolution with balanced Max IT (110 ms), 1 e 5 AGC Target • Note: 10 targets are “untimed”. At 35 K resolution, 1. 3 s cycle time will generate ~10 scans across a 10 sec FWHM peak 45
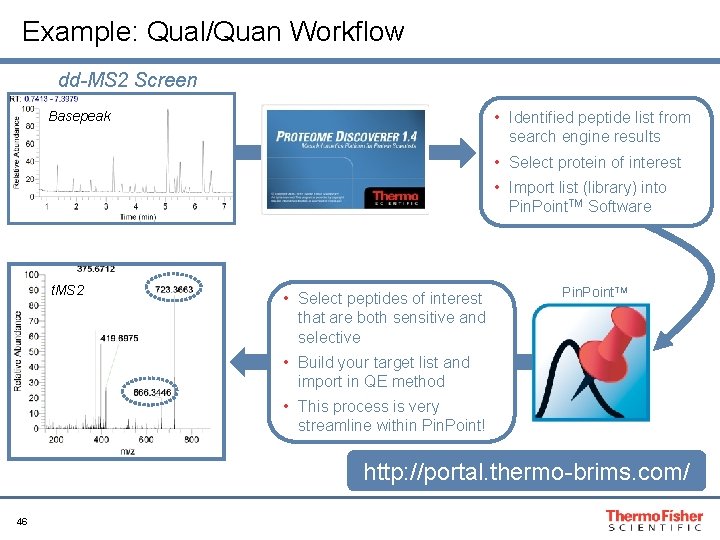
Example: Qual/Quan Workflow dd-MS 2 Screen • Identified peptide list from search engine results Basepeak • Select protein of interest • Import list (library) into Pin. Point. TM Software t. MS 2 • Select peptides of interest that are both sensitive and selective Pin. Point. TM • Build your target list and import in QE method • This process is very streamline within Pin. Point! http: //portal. thermo-brims. com/ 46
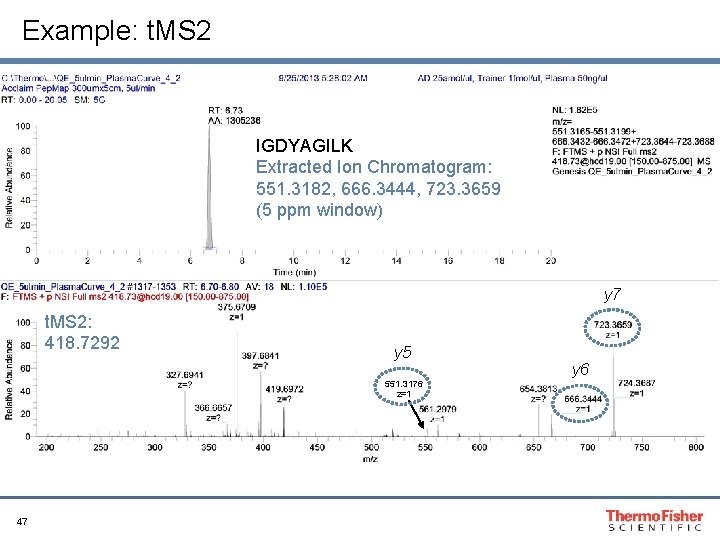
Example: t. MS 2 IGDYAGILK Extracted Ion Chromatogram: 551. 3182, 666. 3444, 723. 3659 (5 ppm window) y 7 t. MS 2: 418. 7292 y 5 551. 3176 z=1 47 y 6
Pin. Point: Detailed Data Analysis 48
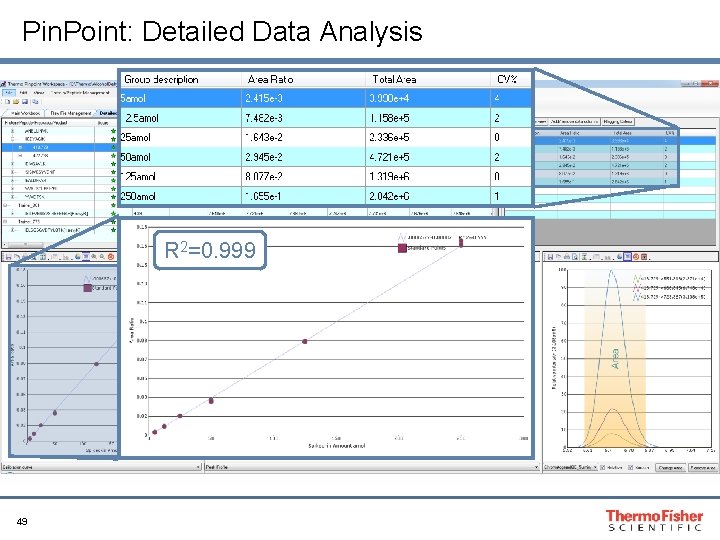
Pin. Point: Detailed Data Analysis R 2=0. 999 49
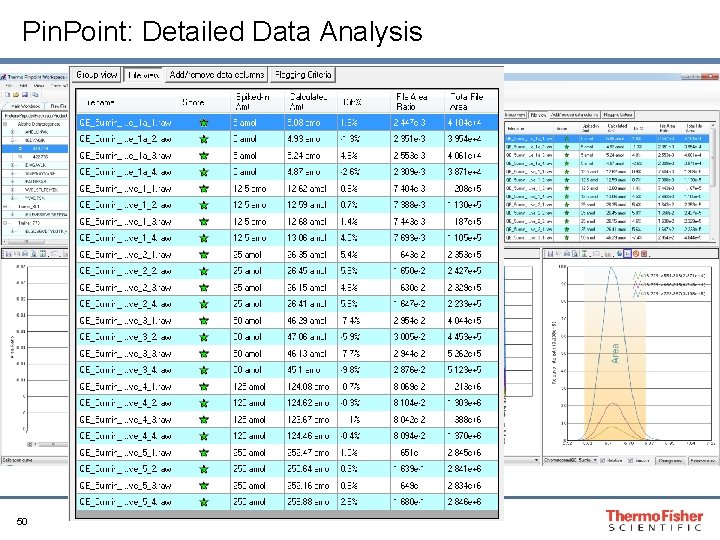
Pin. Point: Detailed Data Analysis 50

Guide to “How to Design an Optimized Quan Method? ” and Troubleshooting Tips
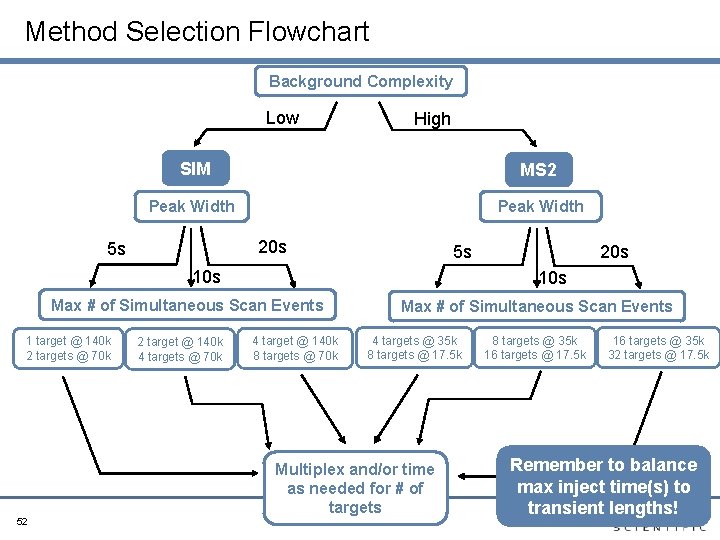
Method Selection Flowchart Background Complexity Low High SIM MS 2 Peak Width 20 s 5 s 5 s 10 s Max # of Simultaneous Scan Events 1 target @ 140 k 2 targets @ 70 k 52 2 target @ 140 k 4 targets @ 70 k 20 s 4 target @ 140 k 8 targets @ 70 k Max # of Simultaneous Scan Events 4 targets @ 35 k 8 targets @ 17. 5 k Multiplex and/or time as needed for # of targets 8 targets @ 35 k 16 targets @ 17. 5 k 16 targets @ 35 k 32 targets @ 17. 5 k Remember to balance max inject time(s) to transient lengths!
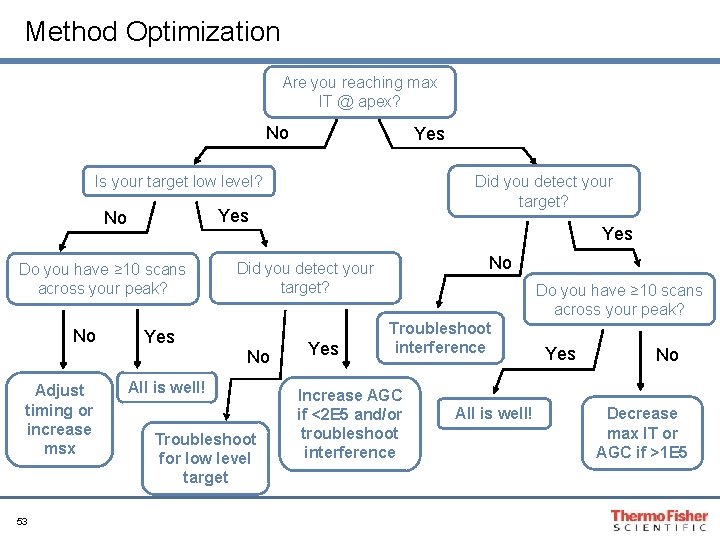
Method Optimization Are you reaching max IT @ apex? No Yes Is your target low level? Yes No Do you have ≥ 10 scans across your peak? No Adjust timing or increase msx 53 Did you detect your target? Yes No Did you detect your target? No All is well! Troubleshoot for low level target Yes Do you have ≥ 10 scans across your peak? Troubleshoot interference Increase AGC if <2 E 5 and/or troubleshoot interference All is well! Yes No Decrease max IT or AGC if >1 E 5

Method Troubleshooting • Interference in your isolation window? • Decrease isolation width • Switch from SIM to t. MS 2 • Adjust gradient conditions • Target is low level and close to limit of detection? • If you are reaching target value, increase target to 2 e 5 • If you are not reaching target, then increase max IT as high as peak width allows • Play close attention to the timing of your targets. If you have very reproducible RT, then try to narrow your time segments which can allow you to increase you max IT. 54
- Slides: 54