How to assure MIAPE compliance of the data







- Slides: 32

How to assure MIAPE compliance of the data using the Proteo. Red MIAPE Extractor tool HUPO-PSI meeting - Liverpool (15 th April 2013) Salvador Martínez-Bartolomé (smartinez@proteored. org) Bioinformatics Support Working Group – Proteo. Red ISCIII National Center for Biotechnology – CNB - CSIC

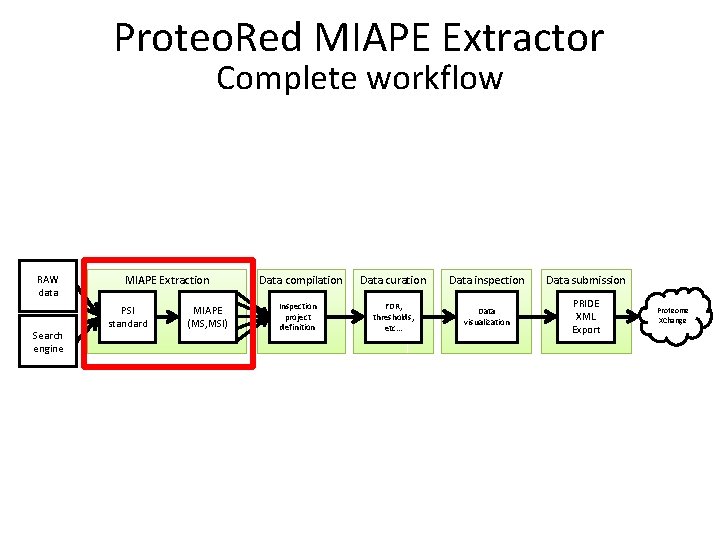
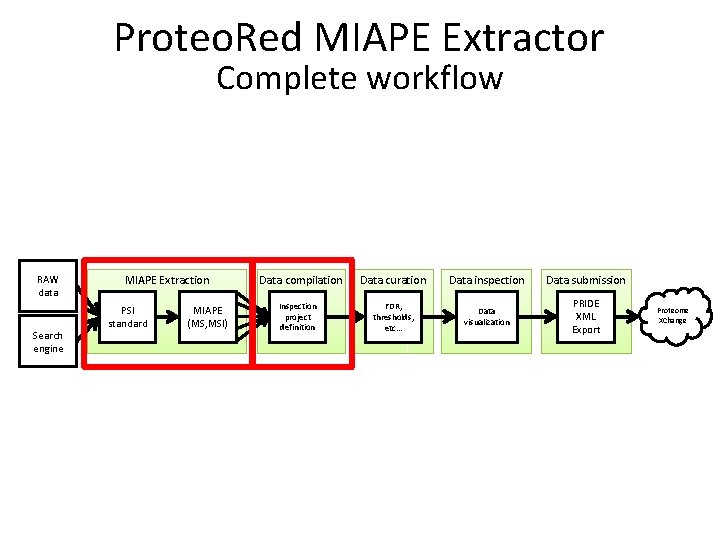
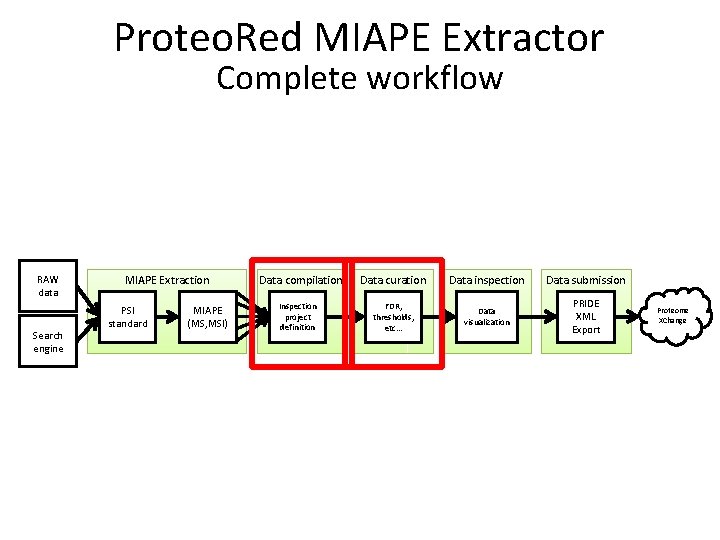
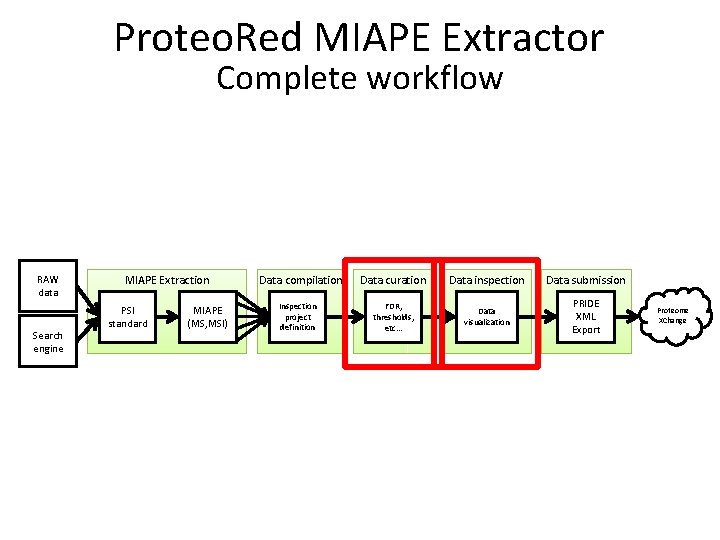
Proteo. Red MIAPE Extractor Complete workflow RAW data Search engine MIAPE Extraction PSI standard MIAPE (MS, MSI) Data compilation Inspection project definition Data curation Data inspection Data submission FDR, thresholds, etc… Data visualization PRIDE XML Export Proteome XChange

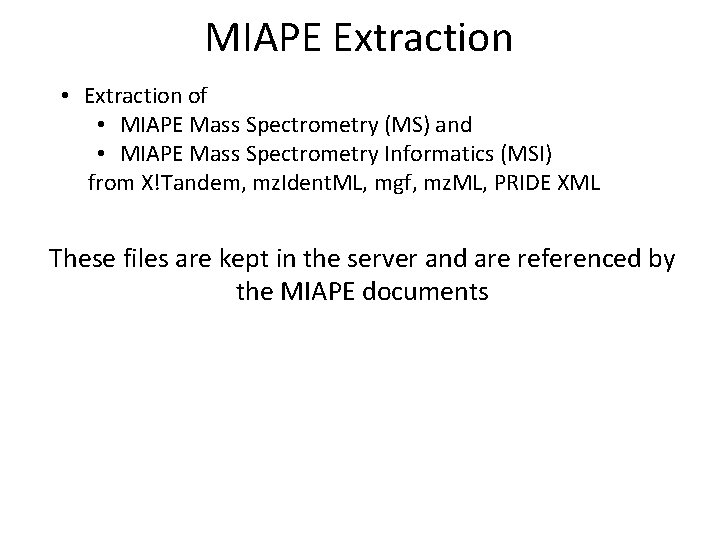
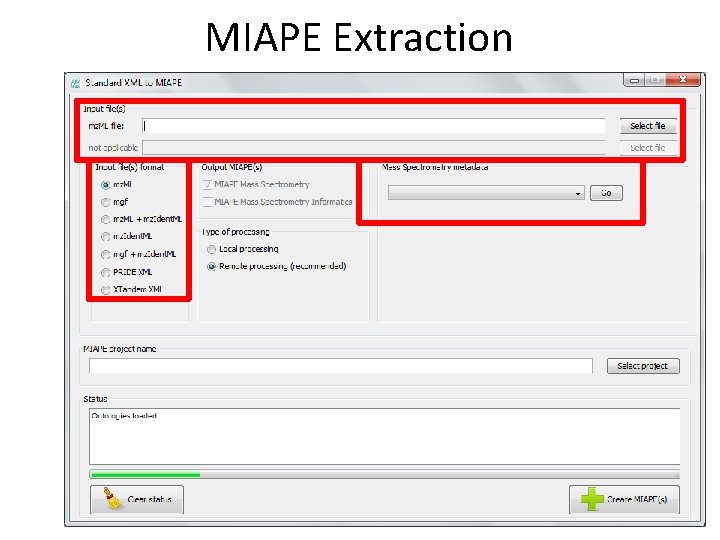
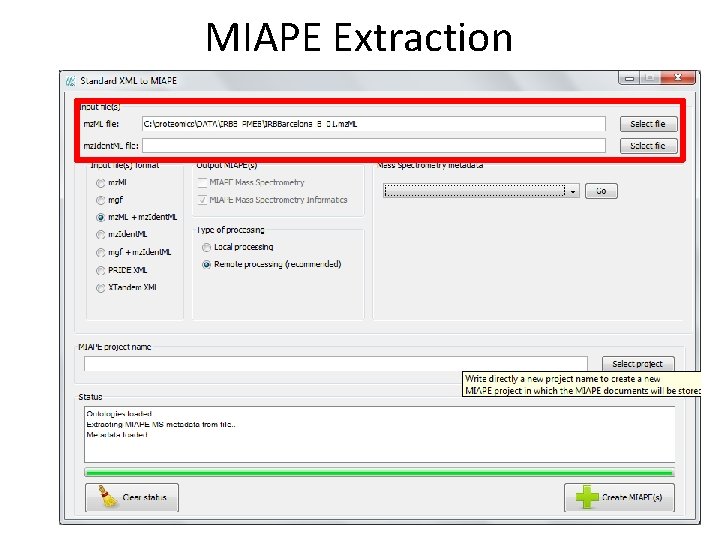
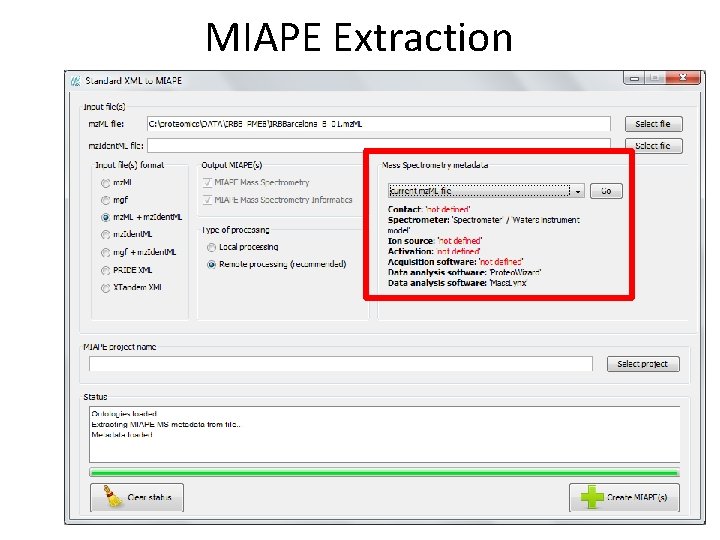
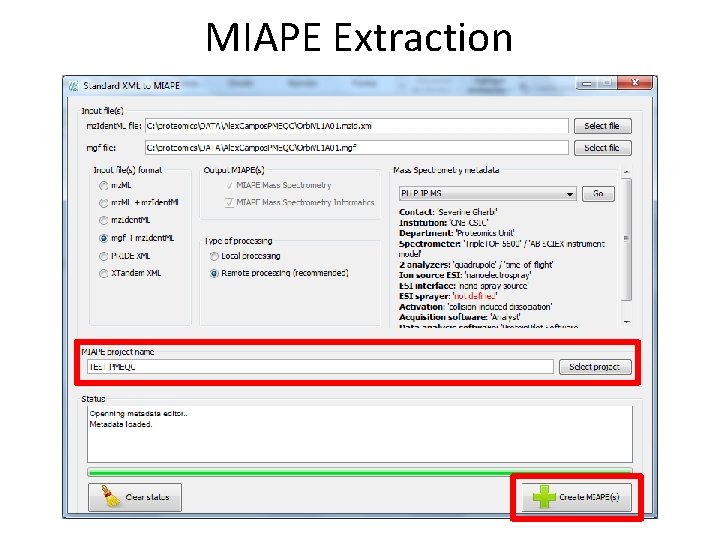
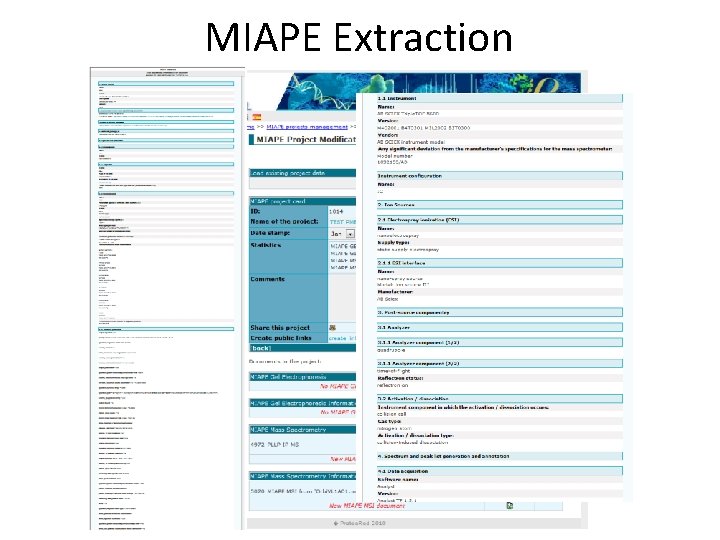
MIAPE Extraction • Extraction of • MIAPE Mass Spectrometry (MS) and • MIAPE Mass Spectrometry Informatics (MSI) from X!Tandem, mz. Ident. ML, mgf, mz. ML, PRIDE XML These files are kept in the server and are referenced by the MIAPE documents

MIAPE Extraction

MIAPE Extraction

MIAPE Extraction

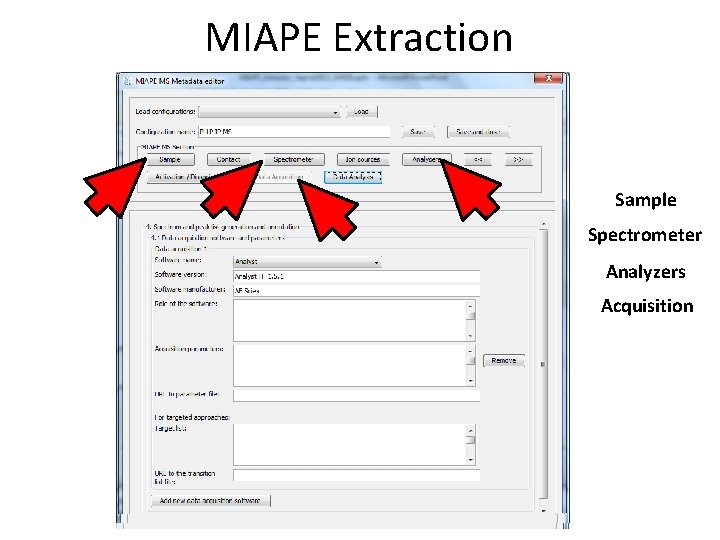
MIAPE Extraction Sample Spectrometer Analyzers Acquisition

MIAPE Extraction

MIAPE Extraction

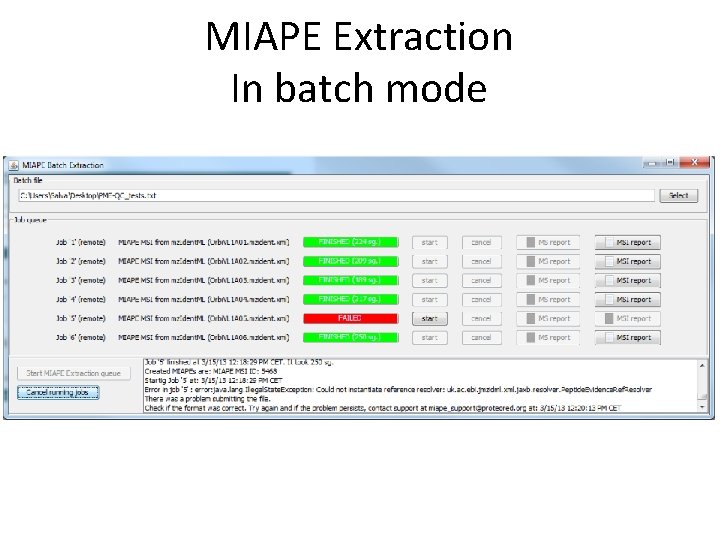
MIAPE Extraction In batch mode

Proteo. Red MIAPE Extractor Complete workflow RAW data Search engine MIAPE Extraction PSI standard MIAPE (MS, MSI) Data compilation Inspection project definition Data curation Data inspection Data submission FDR, thresholds, etc… Data visualization PRIDE XML Export Proteome XChange

Data compilation / aggregation

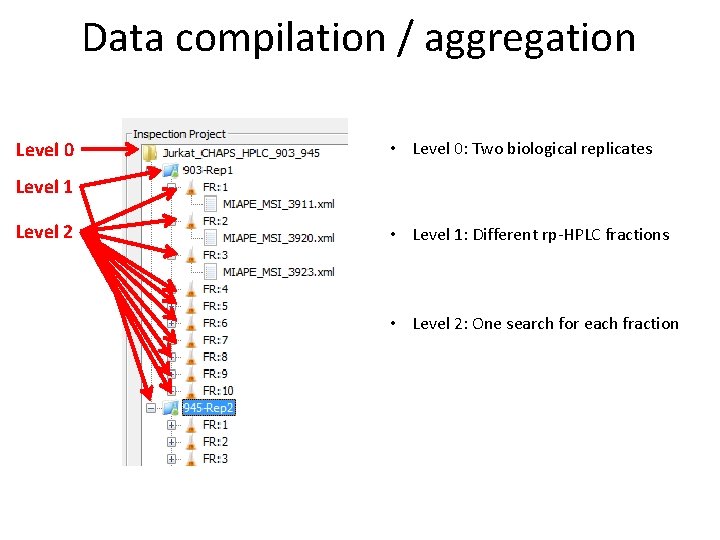
Data compilation / aggregation 1. . n run(s) 1 dataset • User can organize datasets in three different levels: – Level 2: data sets that can be composed by 1. . n runs – Level 1: experiment: fractions, bands – Level 0: experiment list: different experiments, replicates • Define which data is going to be inspected / compared

Data compilation / aggregation Level 0 • Level 0: Two biological replicates Level 1 Level 2 • Level 1: Different rp-HPLC fractions • Level 2: One search for each fraction

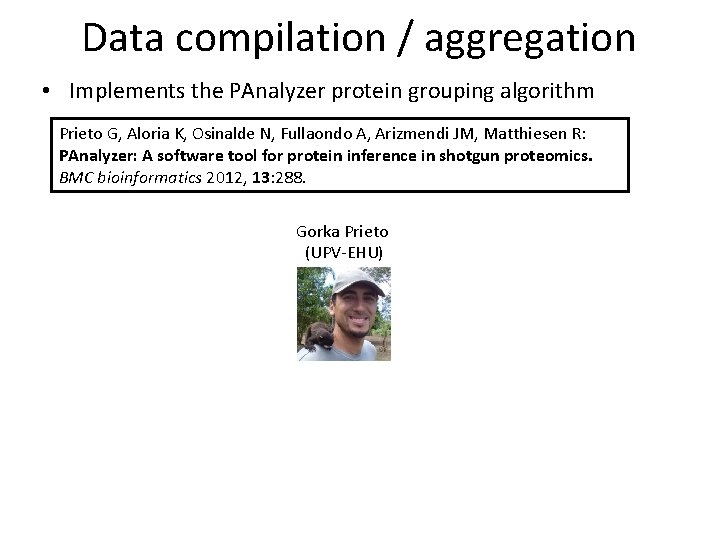
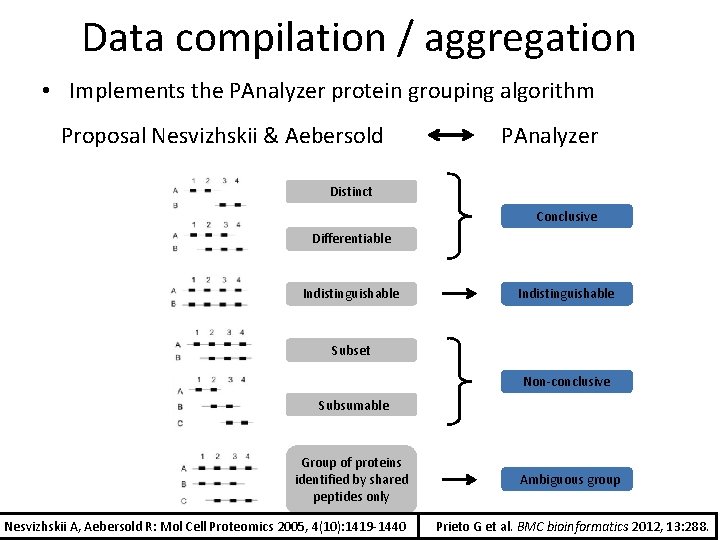
Data compilation / aggregation • Implements the PAnalyzer protein grouping algorithm Prieto G, Aloria K, Osinalde N, Fullaondo A, Arizmendi JM, Matthiesen R: PAnalyzer: A software tool for protein inference in shotgun proteomics. BMC bioinformatics 2012, 13: 288. Gorka Prieto (UPV-EHU)

Data compilation / aggregation • Implements the PAnalyzer protein grouping algorithm Proposal Nesvizhskii & Aebersold PAnalyzer Distinct Conclusive Differentiable Indistinguishable Subset Non-conclusive Subsumable Group of proteins identified by shared peptides only Nesvizhskii A, Aebersold R: Mol Cell Proteomics 2005, 4(10): 1419 -1440 Ambiguous group Prieto G et al. BMC bioinformatics 2012, 13: 288.

Data compilation / aggregation • At experiment level (level 1), protein grouping is made from proteins and peptides coming from all fractions: • • Proteins and peptides are collected from fractions Then, PAnalyzer algorithm is run over them

Proteo. Red MIAPE Extractor Complete workflow RAW data Search engine MIAPE Extraction PSI standard MIAPE (MS, MSI) Data compilation Inspection project definition Data curation Data inspection Data submission FDR, thresholds, etc… Data visualization PRIDE XML Export Proteome XChange

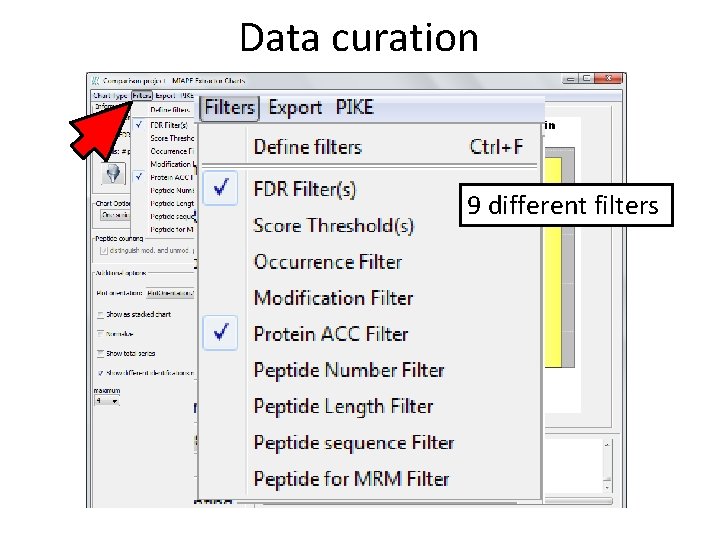
Data curation 9 different filters

Proteo. Red MIAPE Extractor Complete workflow RAW data Search engine MIAPE Extraction PSI standard MIAPE (MS, MSI) Data compilation Inspection project definition Data curation Data inspection Data submission FDR, thresholds, etc… Data visualization PRIDE XML Export Proteome XChange

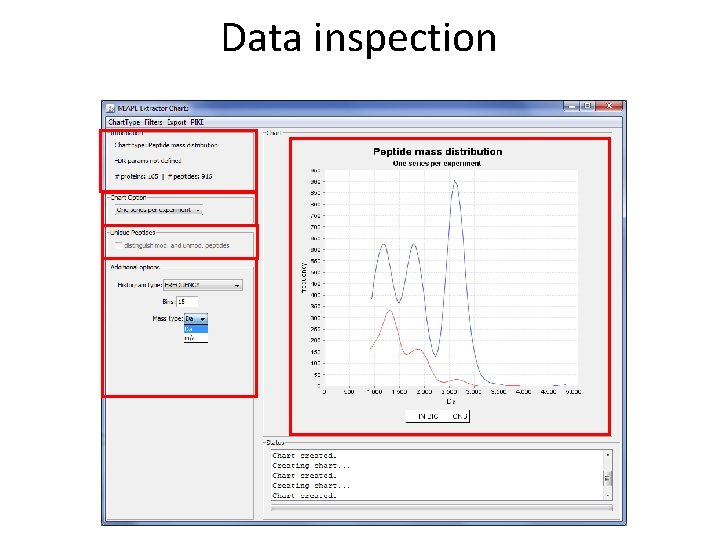
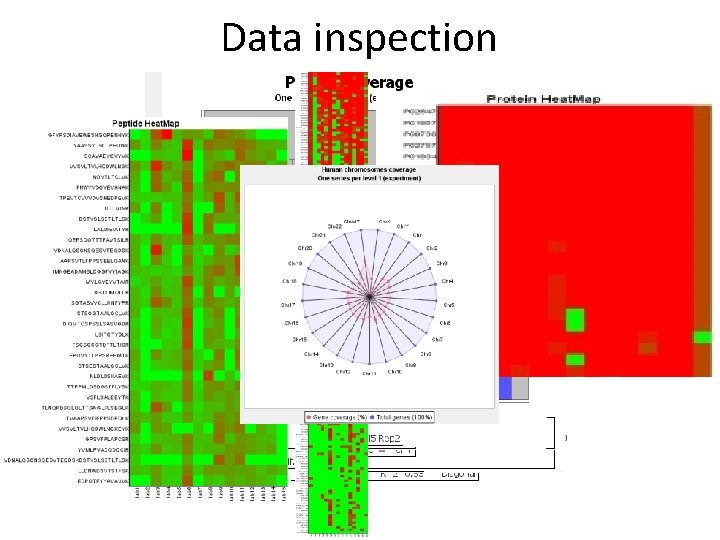
Data inspection

Data inspection 36 different views/plots

Data inspection

Proteo. Red MIAPE Extractor Complete workflow RAW data Search engine MIAPE Extraction PSI standard MIAPE (MS, MSI) Data compilation Inspection project definition Data curation Data inspection Data submission FDR, thresholds, etc… Data visualization PRIDE XML Export Proteome XChange

Data submission • Filtered data will be prepared for a Proteome. Xchange bulk submission • Files attached to the MIAPE reports are downloaded from the MIAPE repository to a local folder: – mz. Ident. ML (attached to the MIAPE MSI) – MGF or mz. ML (attached to the MIAPE MS) • Human readable reports are also generated and downloaded to the local folder

Data submission • A fully MIAPE compliant PRIDE XML file is generated at local folder from each level 1 (normally an experiment/several runs). MTD MTD MTD MTD name email affiliation description keywords type comment species instrument modification pubmed pride_login Salvador Martinez-Bartolome smartinez@proteored. org CNB-CSIC Proteomics Unit Project short description HUPO-PSI, Proteo. Red, MIAPE SUPPORTED This is a comment from the test [NEWT, 9606, Human, ] [MS, MS: 1000447, LTQ, ] [UNIMOD, UNIMOD: 1, Acetyl, ] [UNIMOD, UNIMOD: 4, Carbamidomethyl, ] [UNIMOD, UNIMOD: 35, Oxidation, ] 1234234 smartinez • RAW data files can be added and related to the existing files • A Proteome. Xchange summary file is generated containing the list of files to submit and their relationships FMH FME FME FME FME FME FME FME FME FME FME FME file_id file_type file_path file_mapping 1 result C: proteome. Xchange. Exporter. Test964 -HPLC-RF CNB Jurkat_PRIDE. xml 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 2 raw c: proteome. Xchange. Exporter. TestRAW_File_1_fake. xml 3 raw c: proteome. Xchange. Exporter. TestRAW_File_2_fake. xml 4 raw c: proteome. Xchange. Exporter. TestRAW_File_3_fake. xml 5 raw c: proteome. Xchange. Exporter. TestRAW_File_4_fake. xml 6 raw c: proteome. Xchange. Exporter. TestRAW_File_5_fake. xml 7 raw c: proteome. Xchange. Exporter. TestRAW_File_6_fake. xml 8 raw c: proteome. Xchange. Exporter. TestRAW_File_7_fake. xml 9 raw c: proteome. Xchange. Exporter. TestRAW_File_8_fake. xml 10 raw c: proteome. Xchange. Exporter. TestRAW_File_9_fake. xml 11 raw c: proteome. Xchange. Exporter. TestRAW_File_10_fake. xml 12 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 1. mgf. gz 13 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 2. mgf. gz 14 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 3. mgf. gz 15 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 4. mgf. gz 16 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 5. mgf. gz 17 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 6. mgf. gz 18 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 7. mgf. gz 19 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 8. mgf. gz 20 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 9. mgf. gz 21 peak c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 10. mgf. gz 22 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 1_MIAPE_MS_report. html 23 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 2_MIAPE_MS_report. html 24 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 3_MIAPE_MS_report. html 25 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 4_MIAPE_MS_report. html 26 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 5_MIAPE_MS_report. html 27 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 6_MIAPE_MS_report. html 28 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 7_MIAPE_MS_report. html 29 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 8_MIAPE_MS_report. html 30 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 9_MIAPE_MS_report. html 31 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 10_MIAPE_MS_report. html 32 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 1_MIAPE_MSI_report. html 33 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 2_MIAPE_MSI_report. html 34 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 3_MIAPE_MSI_report. html 35 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 4_MIAPE_MSI_report. html 36 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 5_MIAPE_MSI_report. html 37 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 6_MIAPE_MSI_report. html 38 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 7_MIAPE_MSI_report. html 39 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 8_MIAPE_MSI_report. html 40 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 9_MIAPE_MSI_report. html 41 other c: proteome. Xchange. Exporter. TestHPP_CHAPS_FR 10_MIAPE_MSI_report. html 1, 2 1, 3 1, 4 1, 5 1, 6 1, 7 1, 8 1, 9 1, 10 1, 11 1, 2, 12 1, 3, 13 1, 4, 14 1, 5, 15 1, 6, 16 1, 7, 17 1, 8, 18 1, 9, 19 1, 10, 20 1, 11, 21

Data submission

Data submission

Data submission

Data submission Proteome. Xchange submission tool http: //www. proteomexchange. org/submission

The Proteo. Red MIAPE repository 10. 063. 199 peptides (784. 276 distinct) 1. 640. 782 proteins (35. 351 distinct)

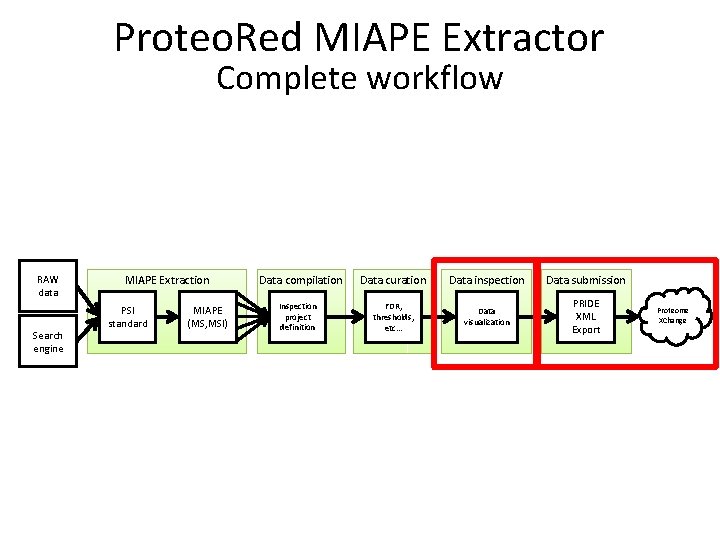
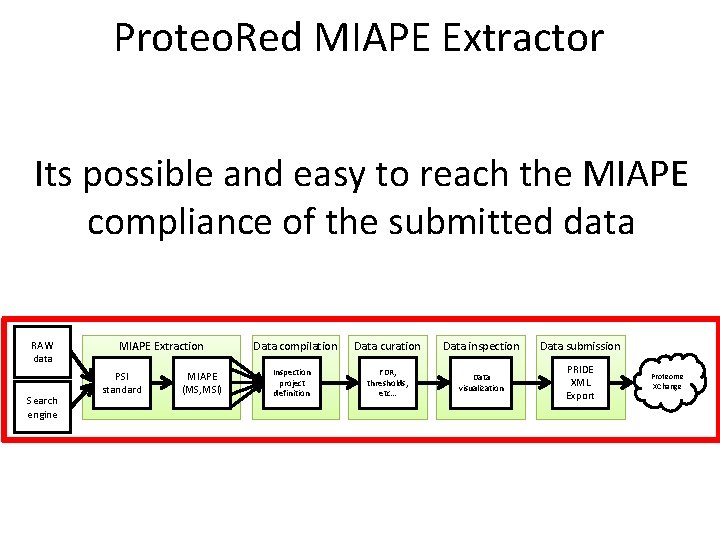
Proteo. Red MIAPE Extractor Its possible and easy to reach the MIAPE compliance of the submitted data RAW data Search engine MIAPE Extraction PSI standard MIAPE (MS, MSI) Data compilation Inspection project definition Data curation Data inspection Data submission FDR, thresholds, etc… Data visualization PRIDE XML Export Proteome XChange
 Miape
Miape Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Lp html
Lp html Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Gấu đi như thế nào
Gấu đi như thế nào Thang điểm glasgow
Thang điểm glasgow Hát lên người ơi
Hát lên người ơi Môn thể thao bắt đầu bằng chữ f
Môn thể thao bắt đầu bằng chữ f Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Công của trọng lực
Công của trọng lực Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Mật thư anh em như thể tay chân
Mật thư anh em như thể tay chân Làm thế nào để 102-1=99
Làm thế nào để 102-1=99 Phản ứng thế ankan
Phản ứng thế ankan Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Thể thơ truyền thống
Thể thơ truyền thống Quá trình desamine hóa có thể tạo ra
Quá trình desamine hóa có thể tạo ra Một số thể thơ truyền thống
Một số thể thơ truyền thống Cái miệng nó xinh thế chỉ nói điều hay thôi
Cái miệng nó xinh thế chỉ nói điều hay thôi Vẽ hình chiếu vuông góc của vật thể sau
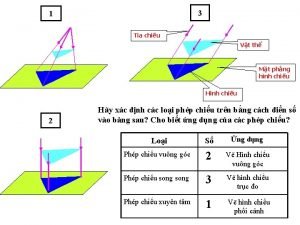
Vẽ hình chiếu vuông góc của vật thể sau Biện pháp chống mỏi cơ
Biện pháp chống mỏi cơ đặc điểm cơ thể của người tối cổ
đặc điểm cơ thể của người tối cổ Ví dụ về giọng cùng tên
Ví dụ về giọng cùng tên Vẽ hình chiếu đứng bằng cạnh của vật thể
Vẽ hình chiếu đứng bằng cạnh của vật thể Phối cảnh
Phối cảnh Thẻ vin
Thẻ vin đại từ thay thế
đại từ thay thế điện thế nghỉ
điện thế nghỉ Tư thế ngồi viết
Tư thế ngồi viết Diễn thế sinh thái là
Diễn thế sinh thái là Dot
Dot