How metal ions control protein structure and dynamics




- Slides: 23

How metal ions control protein structure and dynamics (and biomolecular interactions) A: Calmodulin B: Zinc fingers

Calcium-binding proteins: Calcium regulates protein structure, dynamics and interactions

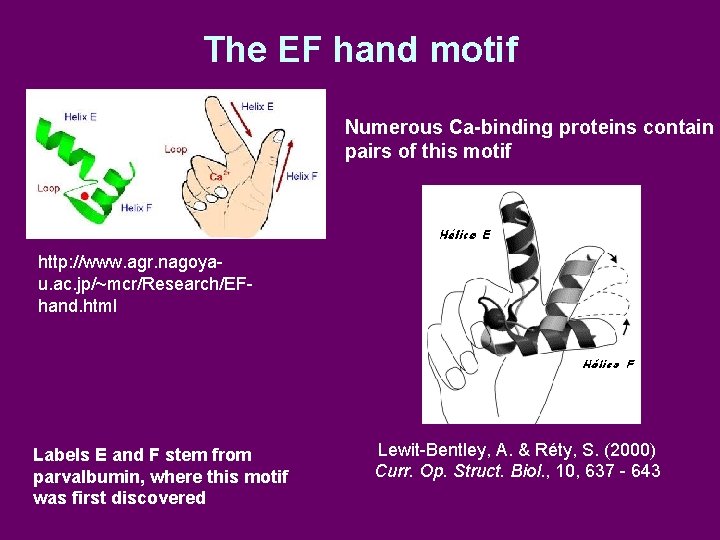
The EF hand motif Numerous Ca-binding proteins contain pairs of this motif http: //www. agr. nagoyau. ac. jp/~mcr/Research/EFhand. html Labels E and F stem from parvalbumin, where this motif was first discovered Lewit-Bentley, A. & Réty, S. (2000) Curr. Op. Struct. Biol. , 10, 637 - 643

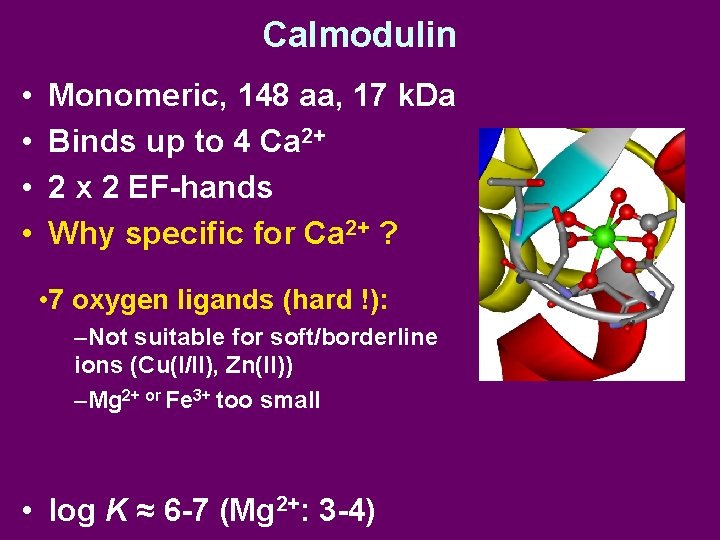
Calmodulin • • Monomeric, 148 aa, 17 k. Da Binds up to 4 Ca 2+ 2 x 2 EF-hands Why specific for Ca 2+ ? • 7 oxygen ligands (hard !): –Not suitable for soft/borderline ions (Cu(I/II), Zn(II)) –Mg 2+ or Fe 3+ too small • log K ≈ 6 -7 (Mg 2+: 3 -4)

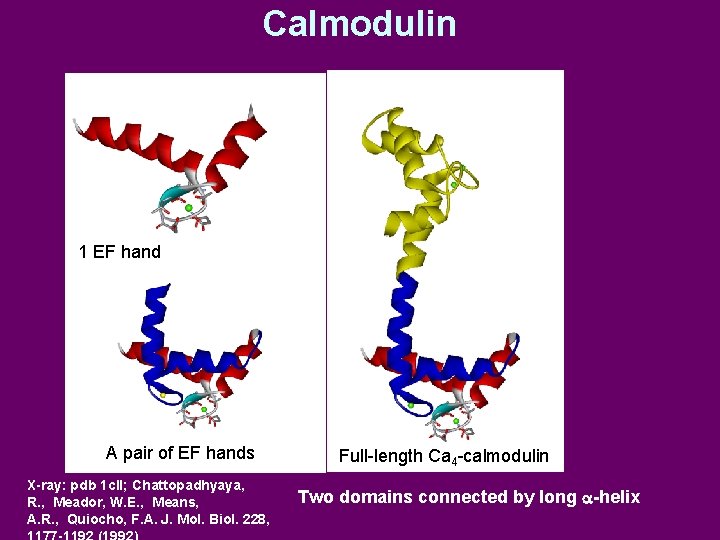
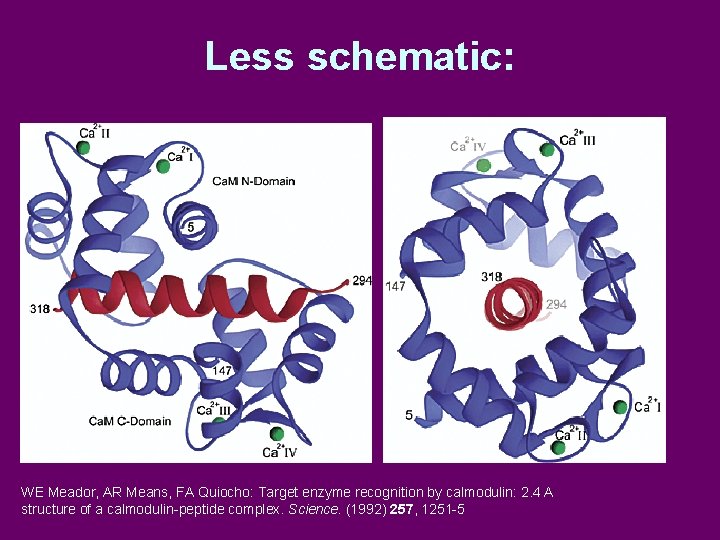
Calmodulin 1 EF hand A pair of EF hands X-ray: pdb 1 cll; Chattopadhyaya, R. , Meador, W. E. , Means, A. R. , Quiocho, F. A. J. Mol. Biol. 228, Full-length Ca 4 -calmodulin Two domains connected by long a-helix

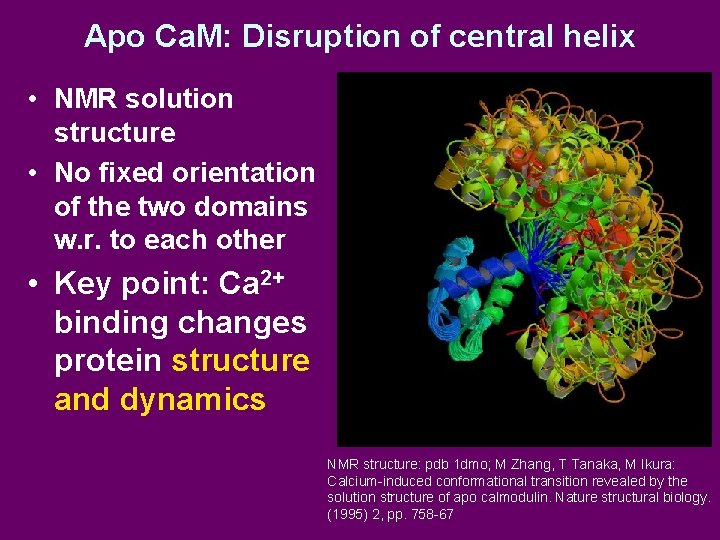
Apo Ca. M: Disruption of central helix • NMR solution structure • No fixed orientation of the two domains w. r. to each other • Key point: Ca 2+ binding changes protein structure and dynamics NMR structure: pdb 1 dmo; M Zhang, T Tanaka, M Ikura: Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nature structural biology. (1995) 2, pp. 758 -67

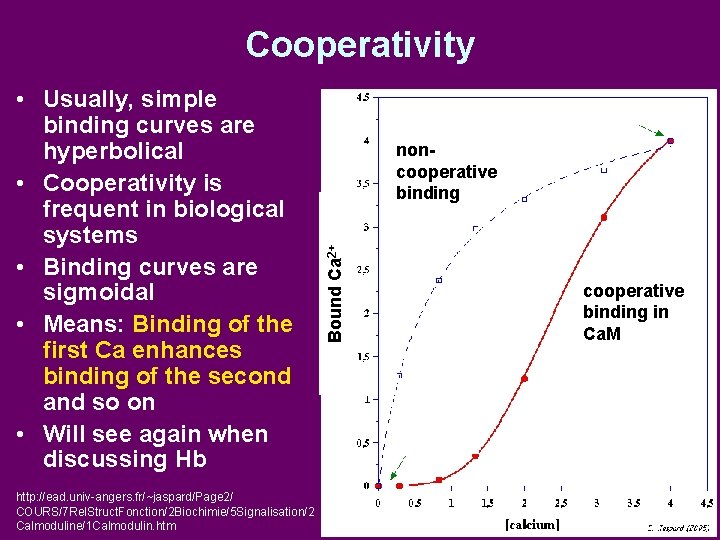
• Usually, simple binding curves are hyperbolical • Cooperativity is frequent in biological systems • Binding curves are sigmoidal • Means: Binding of the first Ca enhances binding of the second and so on • Will see again when discussing Hb http: //ead. univ-angers. fr/~jaspard/Page 2/ COURS/7 Rel. Struct. Fonction/2 Biochimie/5 Signalisation/2 Calmoduline/1 Calmodulin. htm Bound Ca 2+ Cooperativity noncooperative binding in Ca. M

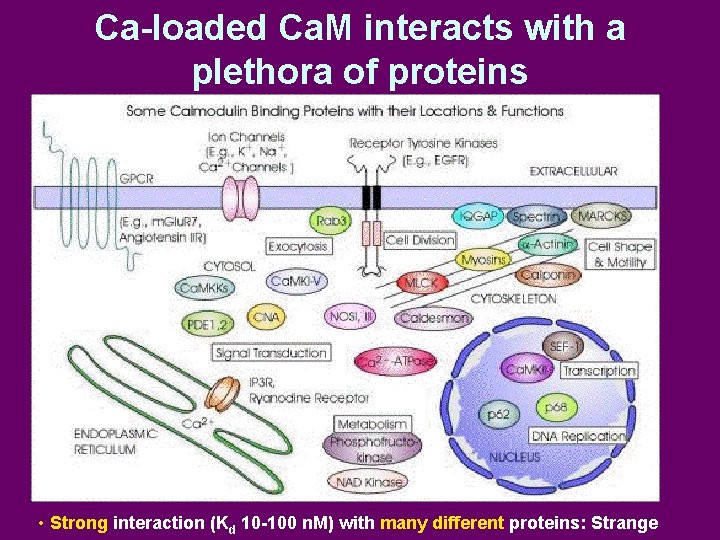
Ca-loaded Ca. M interacts with a plethora of proteins • Strong interaction (Kd 10 -100 n. M) with many different proteins: Strange

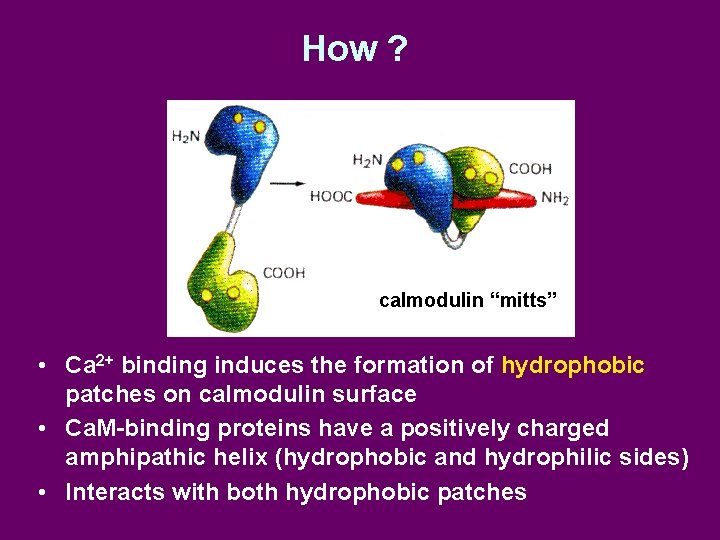
How ? calmodulin “mitts” • Ca 2+ binding induces the formation of hydrophobic patches on calmodulin surface • Ca. M-binding proteins have a positively charged amphipathic helix (hydrophobic and hydrophilic sides) • Interacts with both hydrophobic patches

Less schematic: WE Meador, AR Means, FA Quiocho: Target enzyme recognition by calmodulin: 2. 4 A structure of a calmodulin-peptide complex. Science. (1992) 257, 1251 -5

Summary • Binding of Ca 2+ to calmodulin changes protein structure and dynamics • This change in properties enables Ca. M to bind to a plethora of other proteins, but only in the presence of Ca 2+ (and only Ca 2+) • This is how, by regulation of intracellular Ca 2+ concentrations, the activity of many other proteins can be regulated

Bio-Inorganic Chemistry Lecture 6 b Zinc fingers: Control of protein structure and biomolecular interactions

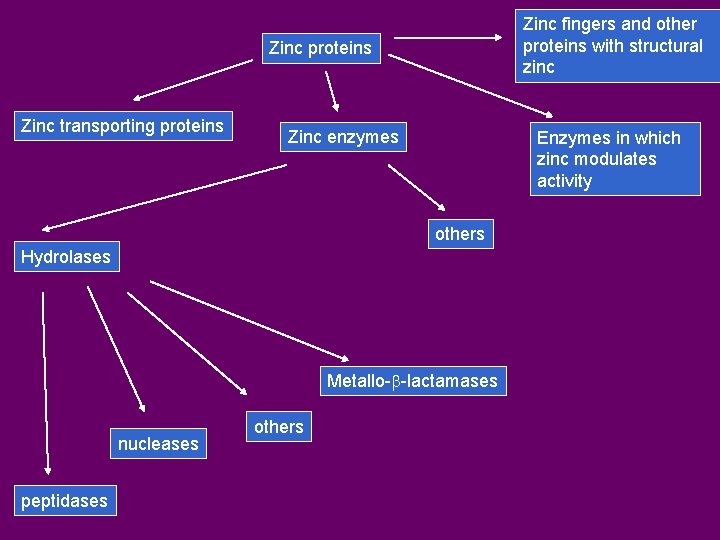
Zinc fingers and other proteins with structural zinc Zinc proteins Zinc transporting proteins Zinc enzymes Enzymes in which zinc modulates activity others Hydrolases Metallo-b-lactamases nucleases peptidases others

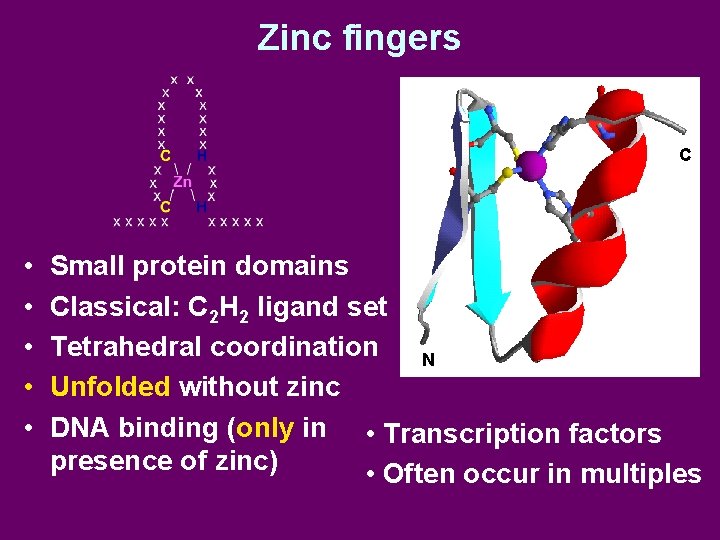
Zinc fingers C • • • Small protein domains Classical: C 2 H 2 ligand set Tetrahedral coordination N Unfolded without zinc DNA binding (only in • Transcription factors presence of zinc) • Often occur in multiples

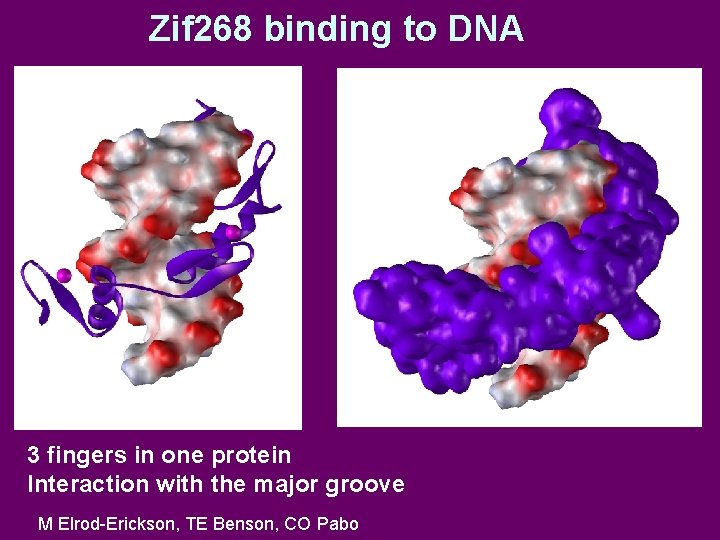
Zif 268 binding to DNA 3 fingers in one protein Interaction with the major groove M Elrod-Erickson, TE Benson, CO Pabo

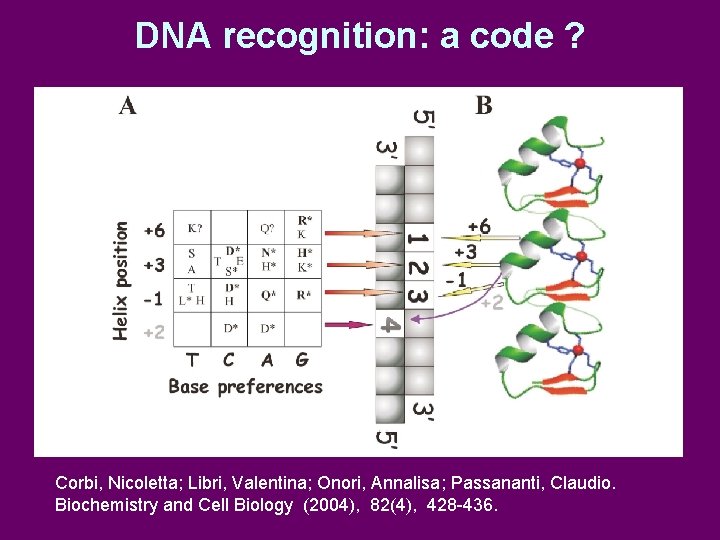
DNA recognition: a code ? Corbi, Nicoletta; Libri, Valentina; Onori, Annalisa; Passananti, Claudio. Biochemistry and Cell Biology (2004), 82(4), 428 -436.

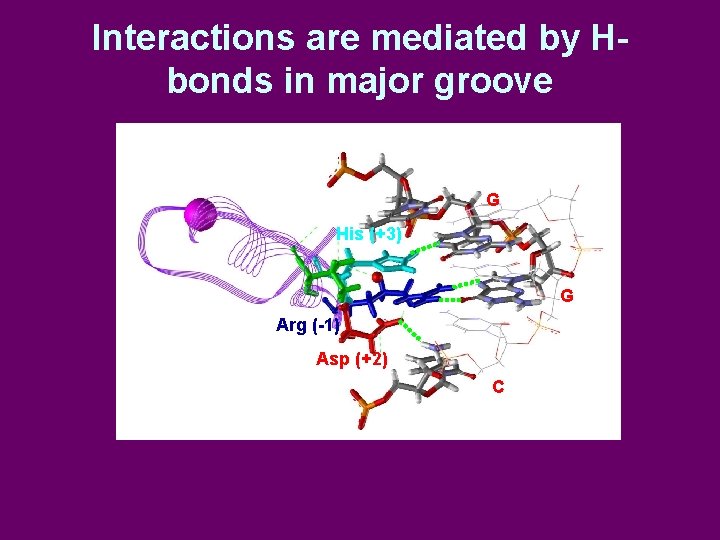
Interactions are mediated by Hbonds in major groove G His (+3) G Arg (-1) Asp (+2) C

Artificial zinc fingers • Based on unravelling the details of zinc finger DNA recognition • Re-design zinc finger proteins for recognition of other DNA sequences • Promising for medical applications: e. g. Gene regulation/targeting, antiviral therapy

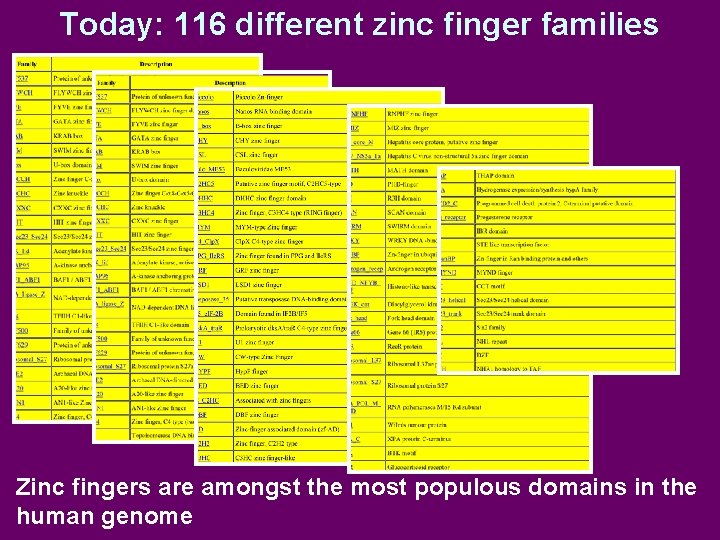
Today: 116 different zinc finger families Zinc fingers are amongst the most populous domains in the human genome

The zinc fingers • Can have His 2 Cys 2, Cys 3 His, or Cys 4 ligand set • Can have 1 or 2 zinc sites • Can be classified by the pattern of ligands in the sequence • E. g. Pattern for GATA-type zinc fingers: • C - x - [DN] - C - x(4, 5) - [ST] - x(2) - W - [HR] - [RK] - x(3) - [GN] - x(3, 4) - C - N - [AS] - C

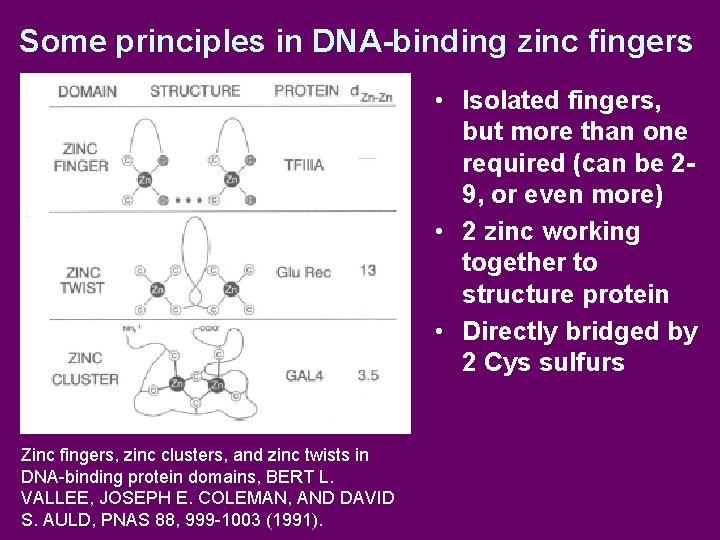
Some principles in DNA-binding zinc fingers • Isolated fingers, but more than one required (can be 29, or even more) • 2 zinc working together to structure protein • Directly bridged by 2 Cys sulfurs Zinc fingers, zinc clusters, and zinc twists in DNA-binding protein domains, BERT L. VALLEE, JOSEPH E. COLEMAN, AND DAVID S. AULD, PNAS 88, 999 -1003 (1991).

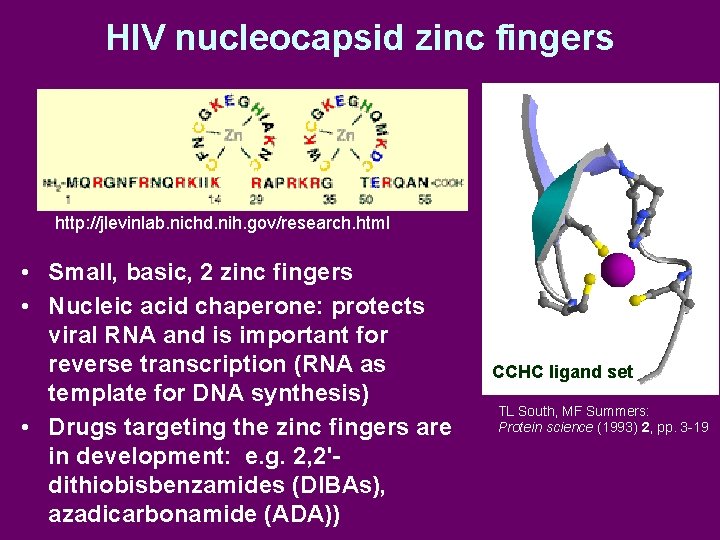
HIV nucleocapsid zinc fingers http: //jlevinlab. nichd. nih. gov/research. html • Small, basic, 2 zinc fingers • Nucleic acid chaperone: protects viral RNA and is important for reverse transcription (RNA as template for DNA synthesis) • Drugs targeting the zinc fingers are in development: e. g. 2, 2'dithiobisbenzamides (DIBAs), azadicarbonamide (ADA)) CCHC ligand set TL South, MF Summers: Protein science (1993) 2, pp. 3 -19

Summary • Zinc fingers are important for mediation of bio-molecular interactions • Cys 2 His 2, Cys 3 His, Cys 4 coordination modes • Unfolded without zinc