How does the cell manufacture these magnificent machines



- Slides: 41

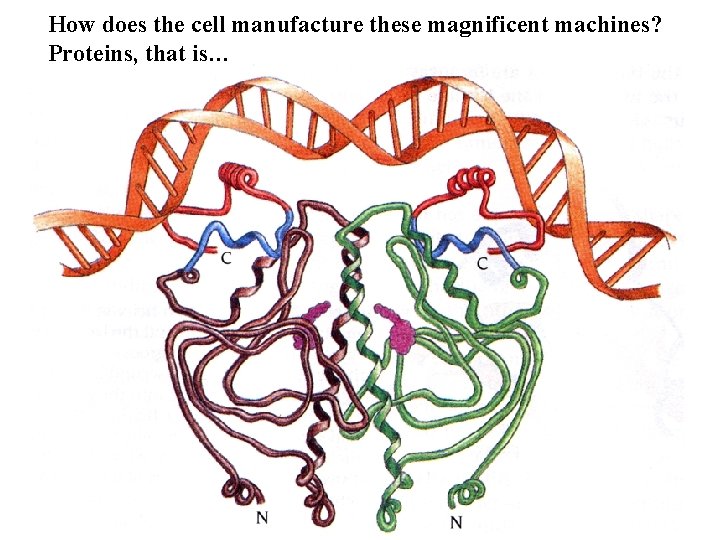
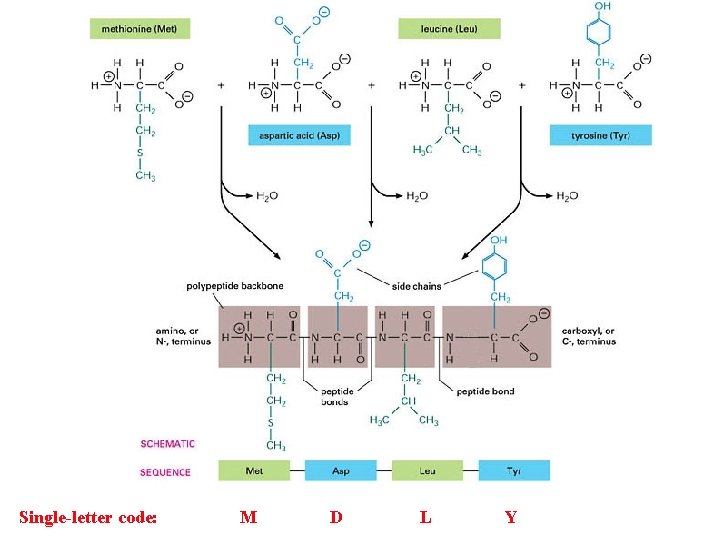
How does the cell manufacture these magnificent machines? Proteins, that is…




Single-letter code: M D L Y

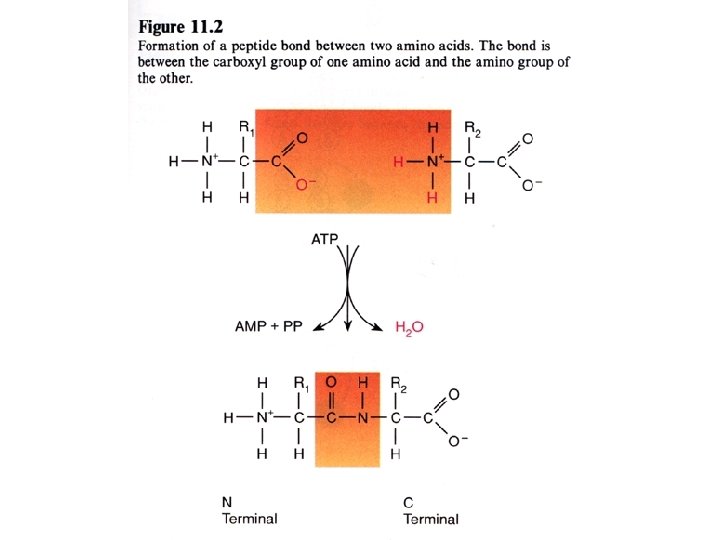
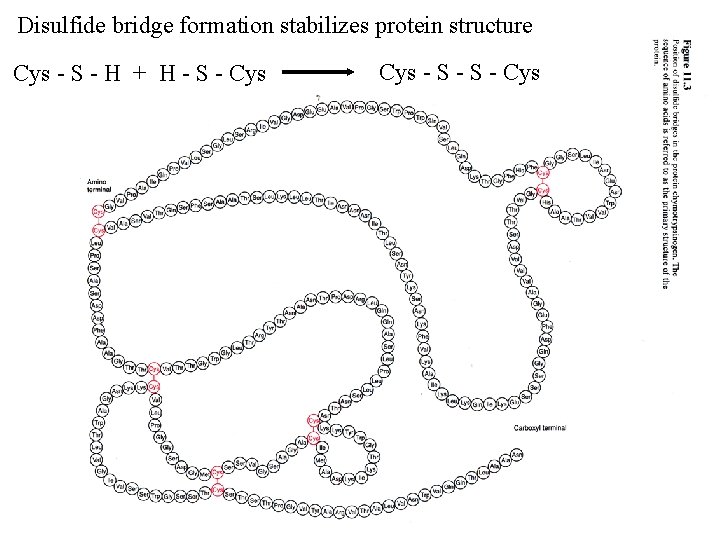
Disulfide bridge formation stabilizes protein structure Cys - S - H + H - S - Cys

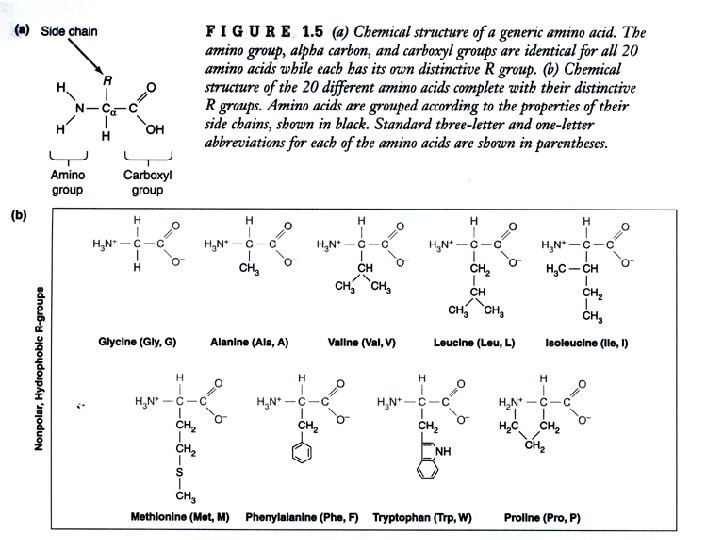
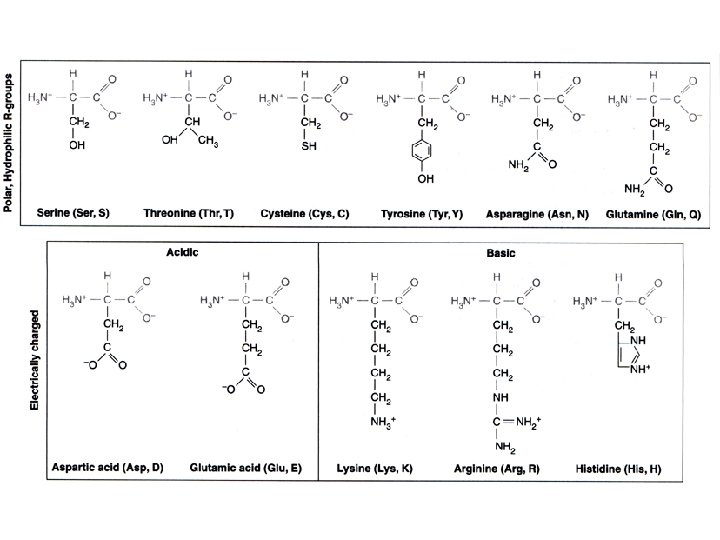
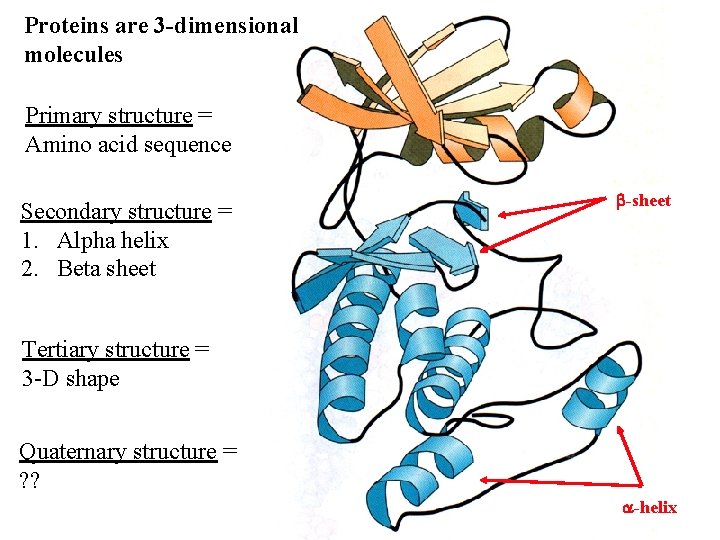
Proteins are 3 -dimensional molecules Primary structure = Amino acid sequence Secondary structure = 1. Alpha helix 2. Beta sheet -sheet Tertiary structure = 3 -D shape Quaternary structure = ? ? -helix

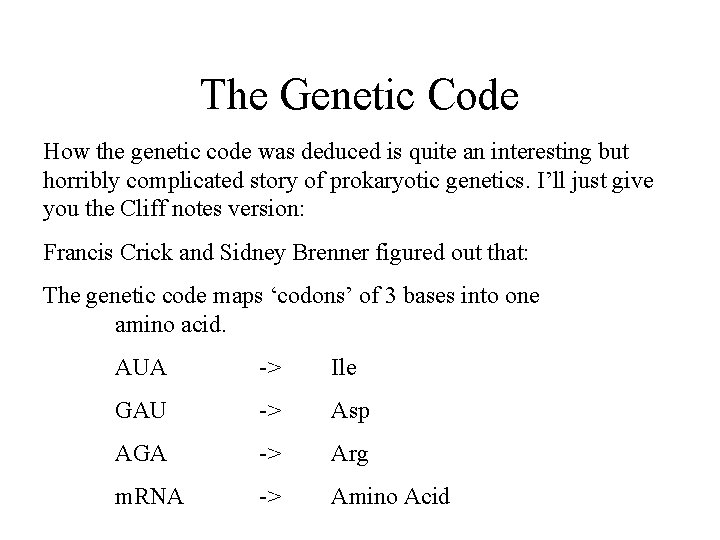
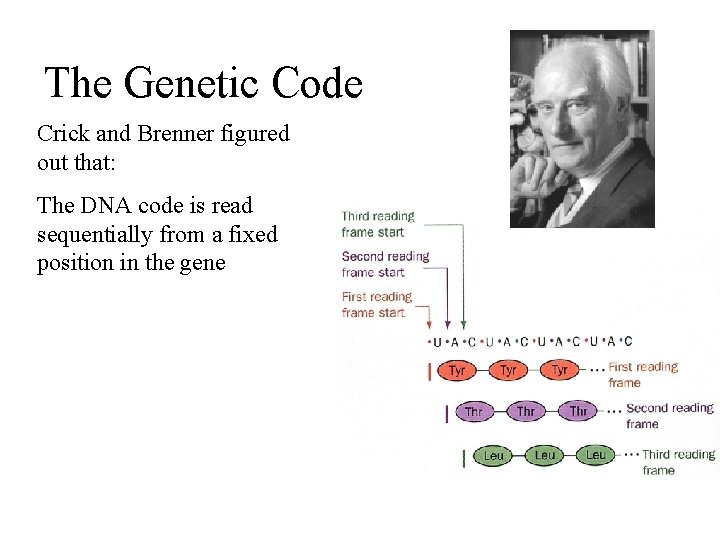
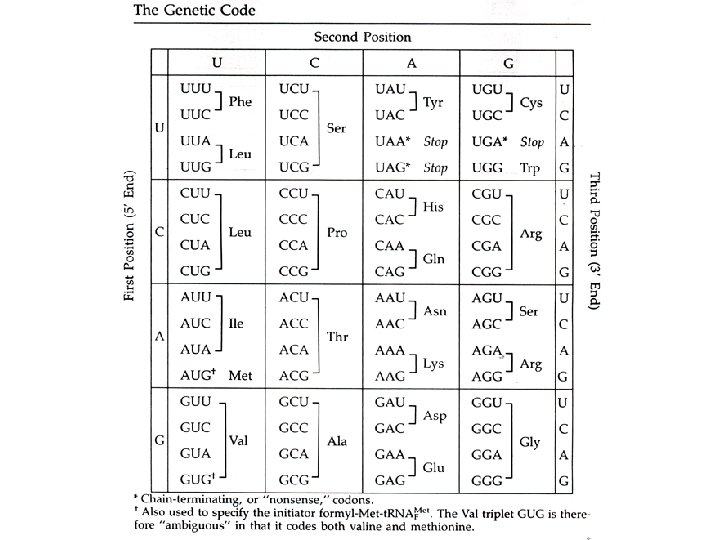
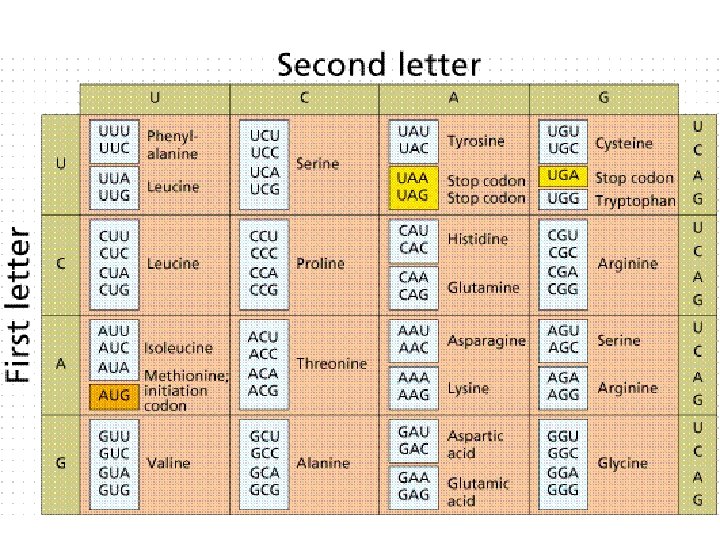
The Genetic Code How the genetic code was deduced is quite an interesting but horribly complicated story of prokaryotic genetics. I’ll just give you the Cliff notes version: Francis Crick and Sidney Brenner figured out that: The genetic code maps ‘codons’ of 3 bases into one amino acid. AUA -> Ile GAU -> Asp AGA -> Arg m. RNA -> Amino Acid

The Genetic Code Crick and Brenner figured out that: The DNA code is read sequentially from a fixed position in the gene

The mechanism and machinery for translating a protein Three components:

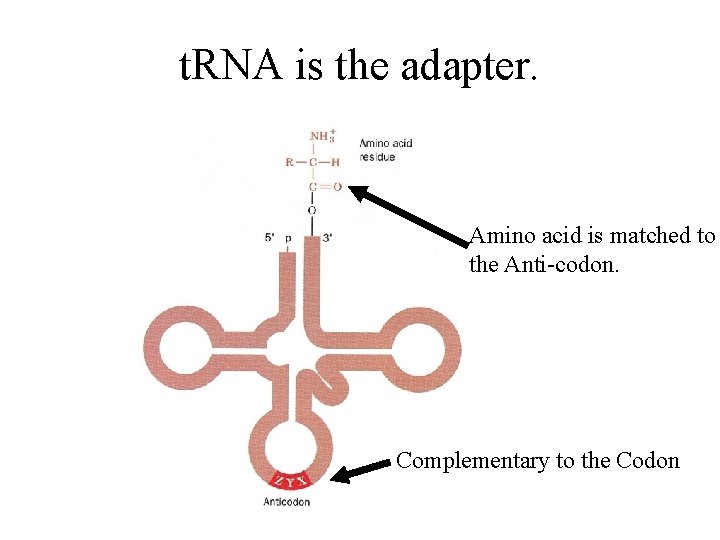
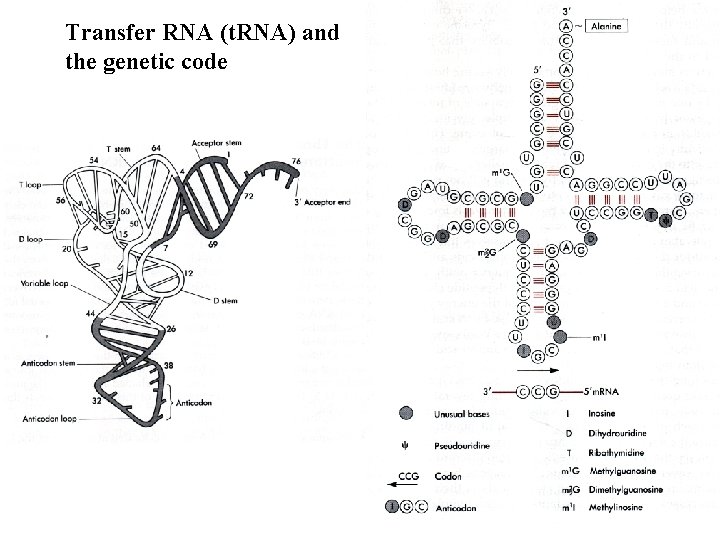
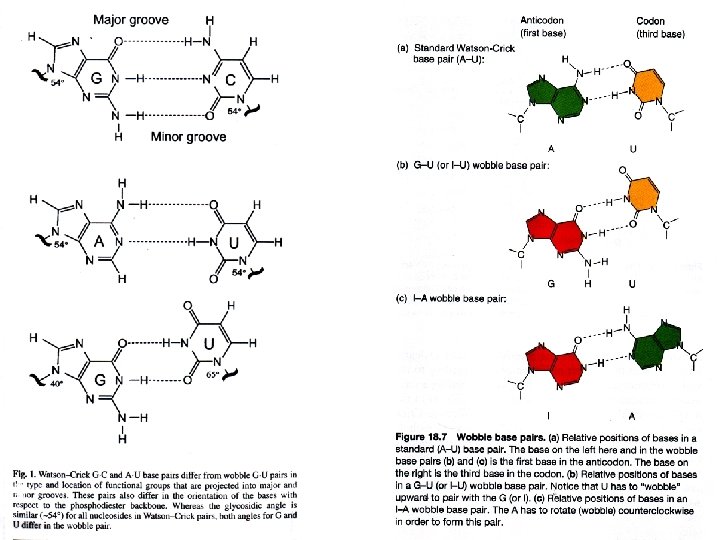
t. RNA is the adapter. Amino acid is matched to the Anti-codon. Complementary to the Codon

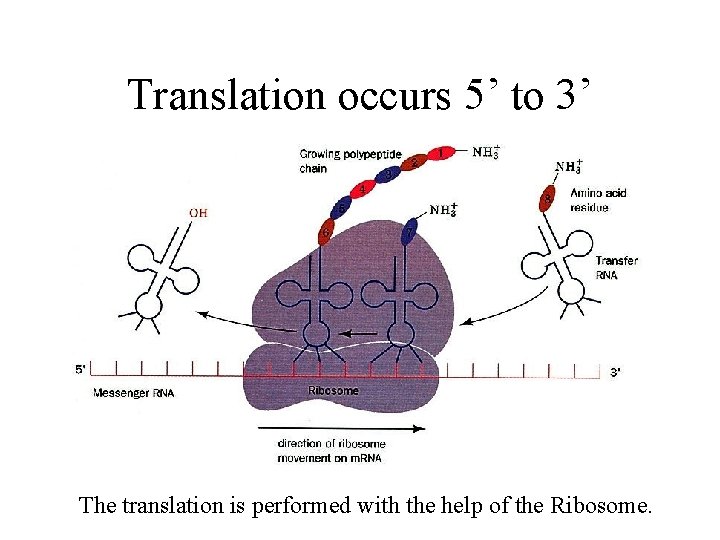
Translation occurs 5’ to 3’ The translation is performed with the help of the Ribosome.

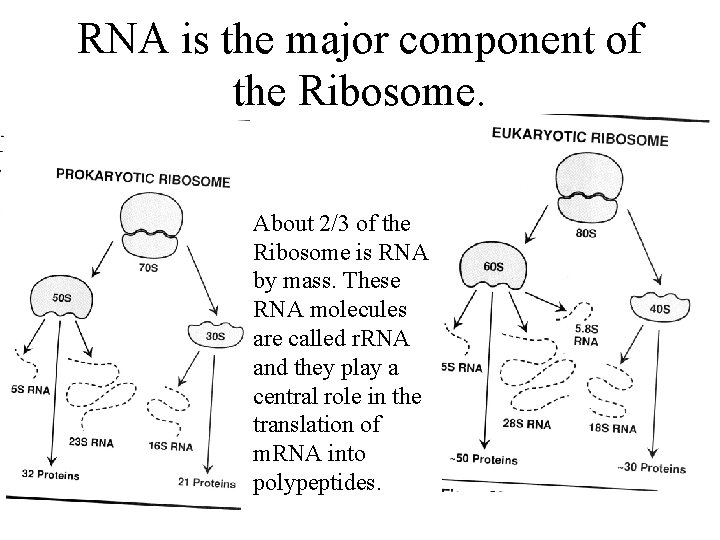
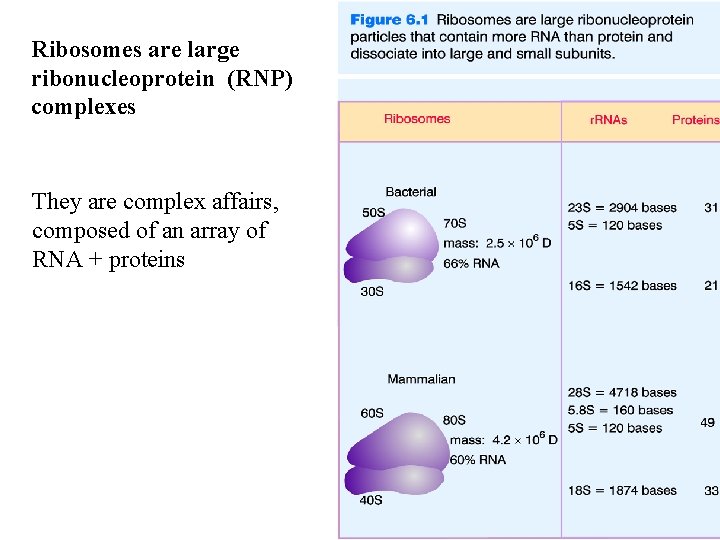
RNA is the major component of the Ribosome. About 2/3 of the Ribosome is RNA by mass. These RNA molecules are called r. RNA and they play a central role in the translation of m. RNA into polypeptides.

m. RNA, r. RNA, t. RNA and protein synthesis In translation, the language of nucleic acids is translated into a new language, that of proteins m. RNA provides the code, in linear digital form, for making a protein t. RNA provides an adaptor that links the code in a polynucleotide chain to amino acids that make up the polypeptide chain r. RNA and ribosomes provide the decoder. Ribosomes bring together m. RNA and t. RNA, and catalyze the translation of an m. RNA into a polypeptide chain. Ribosomes are the site of protein synthesis. Ribosomes create peptide bonds between amino acids to create proteins

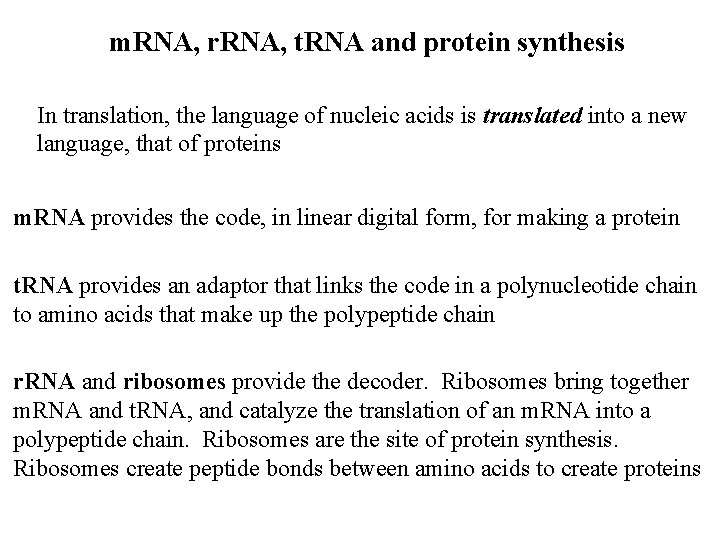
Translation normally occurs on polyribosomes, or polysomes This allows for amplification of the signal from DNA and RNA, i. e. , … One gene copy Hundreds of m. RNAs Thousands of proteins

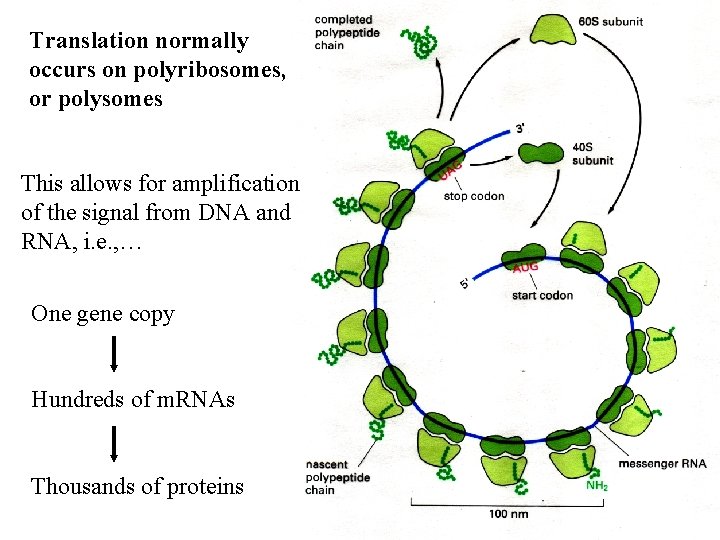
r. RNAs form complex 2 o and 3 o structures that are essential for their function What does this r. RNA molecule look like in three dimensions?

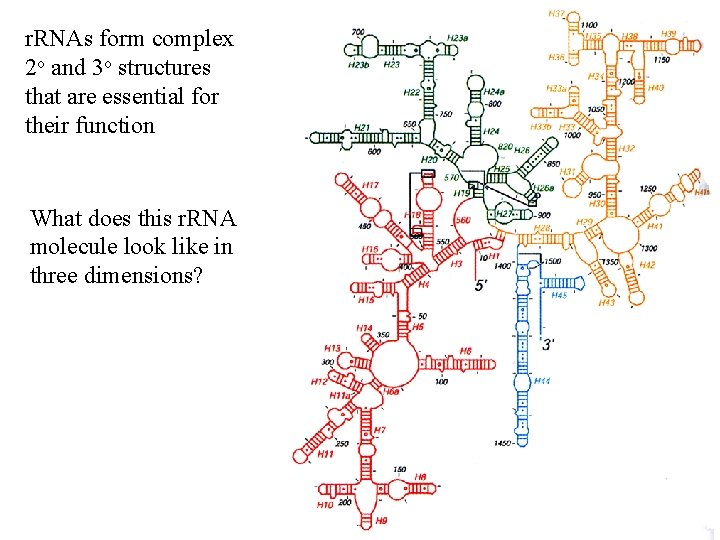
3 -D model of 16 s r. RNA molecule as it folds in the ribosome… …and overlaid with its protein subunits

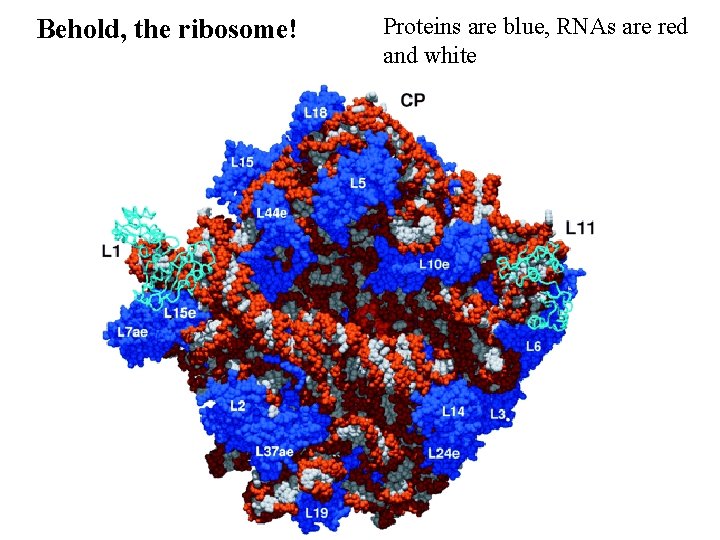
Behold, the ribosome! Proteins are blue, RNAs are red and white

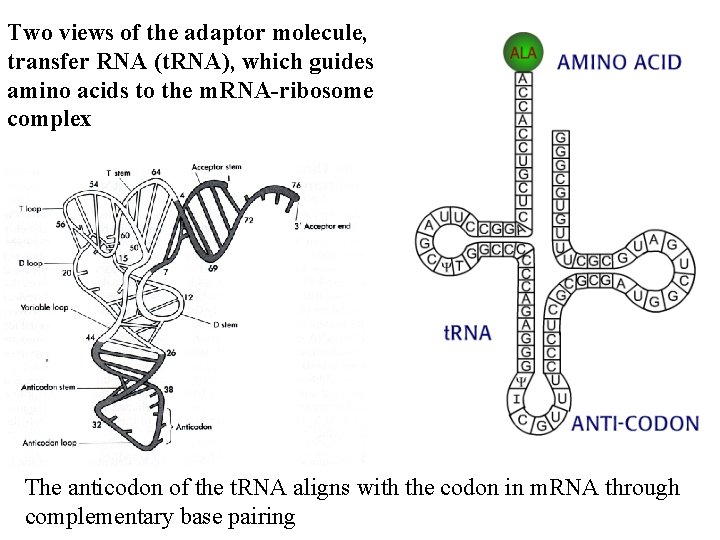
Two views of the adaptor molecule, transfer RNA (t. RNA), which guides amino acids to the m. RNA-ribosome complex The anticodon of the t. RNA aligns with the codon in m. RNA through complementary base pairing

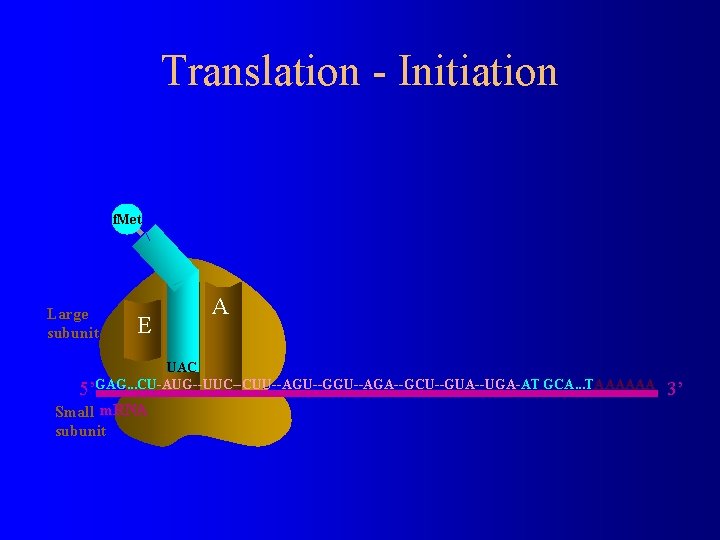
Translation - Initiation f. Met Large subunit E P A UAC 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA Small m. RNA subunit 3’

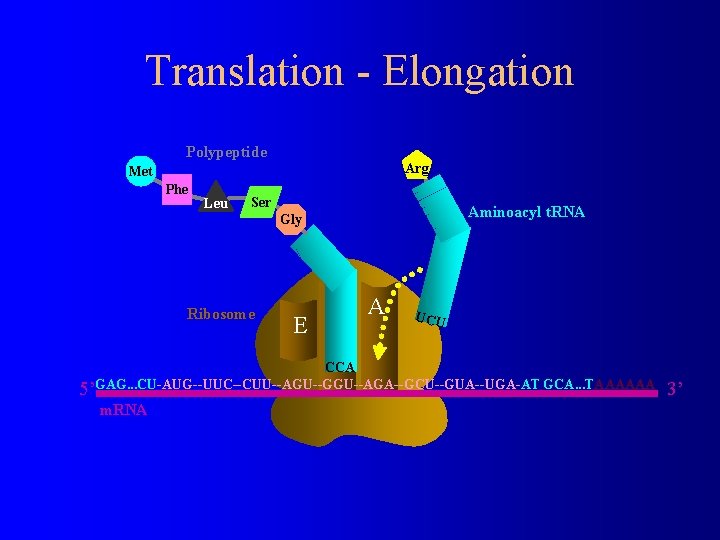
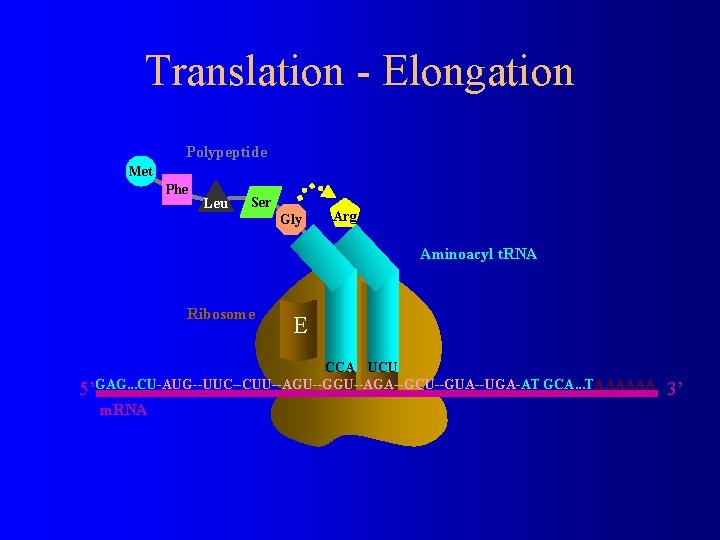
Translation - Elongation Polypeptide Arg Met Phe Leu Ser Aminoacyl t. RNA Gly Ribosome E P A UCU CCA 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’

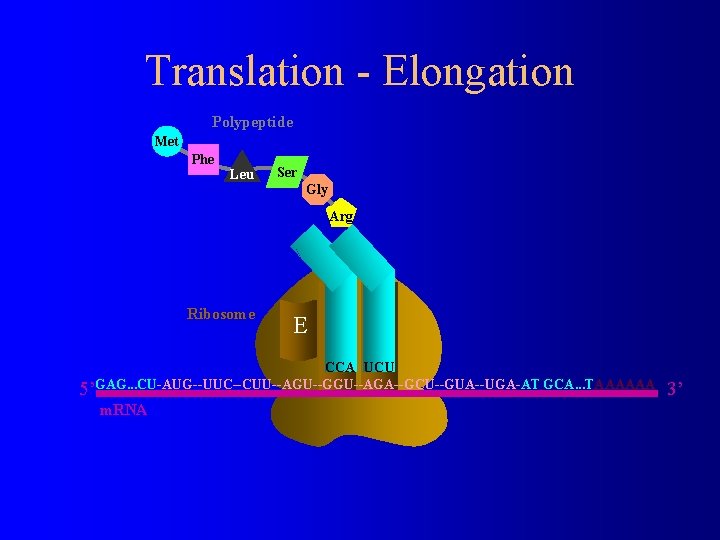
Translation - Elongation Polypeptide Met Phe Leu Ser Gly Arg Aminoacyl t. RNA Ribosome E P A CCA UCU 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’

Translation - Elongation Polypeptide Met Phe Leu Ser Gly Arg Ribosome E P A CCA UCU 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’

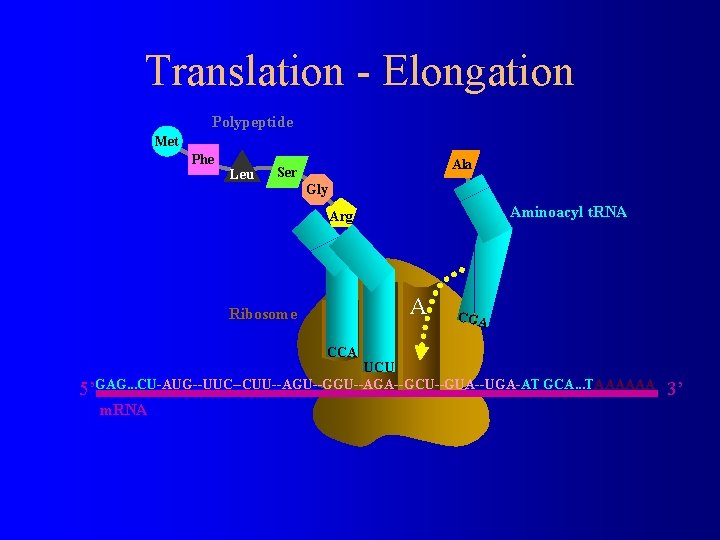
Translation - Elongation Polypeptide Met Phe Leu Ala Ser Gly Aminoacyl t. RNA Arg Ribosome E P A CGA CCA UCU 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’

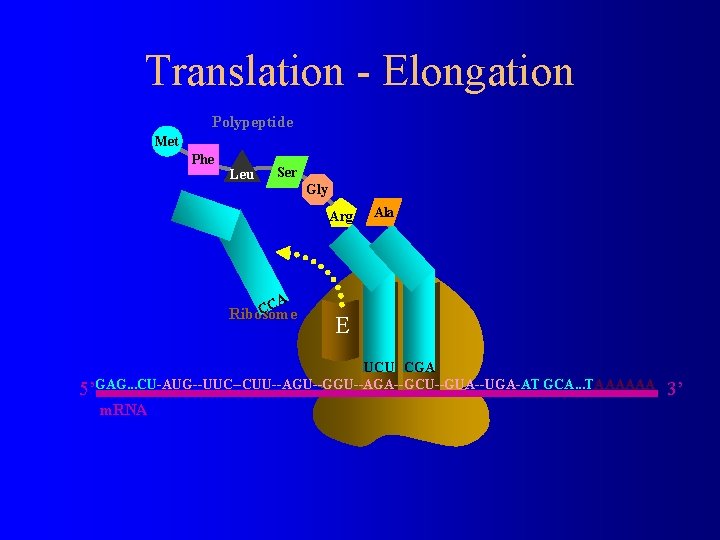
Translation - Elongation Polypeptide Met Phe Leu Ser Gly Arg CA C Ribosome E Ala P A UCU CGA 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’

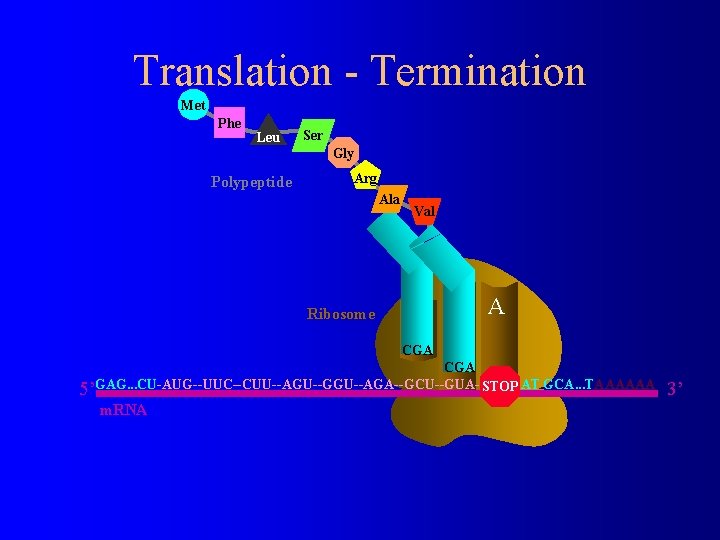
Translation - Termination Met Phe Leu Ser Gly Polypeptide Arg Ala Ribosome Val E P A CGA GCA. . . TAAAAAA STOP 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT m. RNA 3’

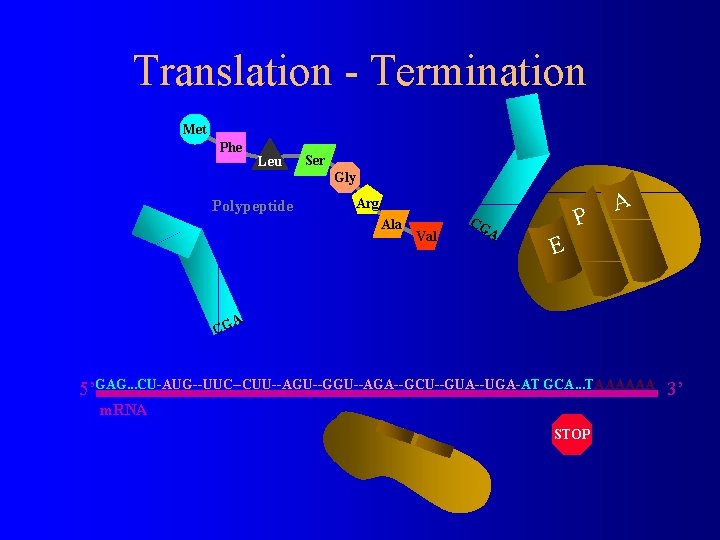
Translation - Termination Met Phe Leu Ser Gly Polypeptide Arg Ala Val CG A P A E A CG 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA 3’ m. RNA STOP

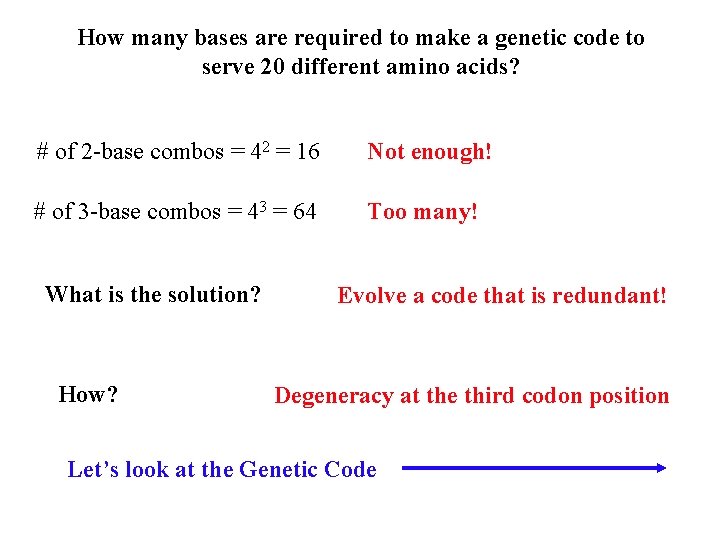
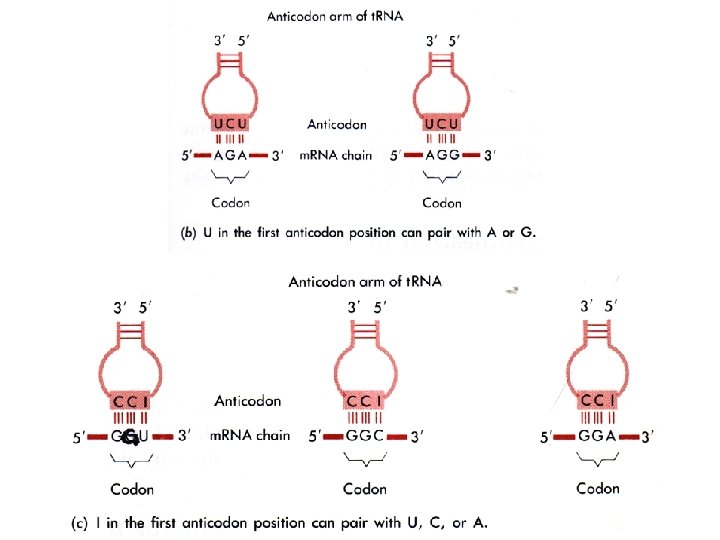
How many bases are required to make a genetic code to serve 20 different amino acids? # of 2 -base combos = 42 = 16 Not enough! # of 3 -base combos = 43 = 64 Too many! What is the solution? How? Evolve a code that is redundant! Degeneracy at the third codon position Let’s look at the Genetic Code



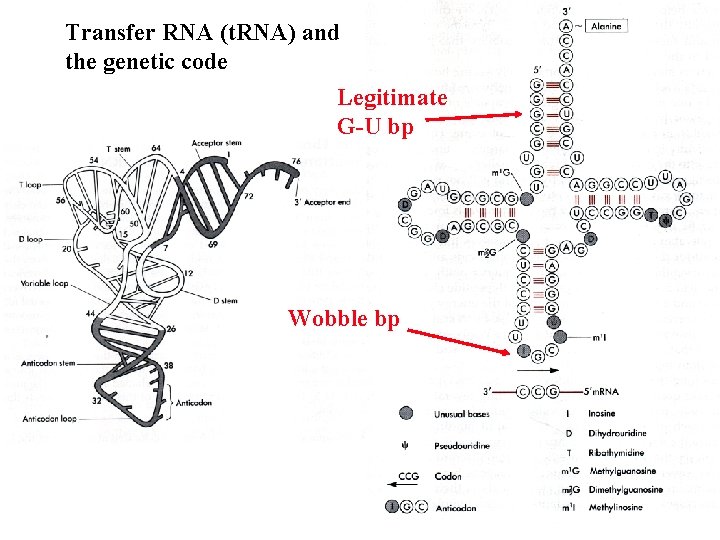
Transfer RNA (t. RNA) and the genetic code

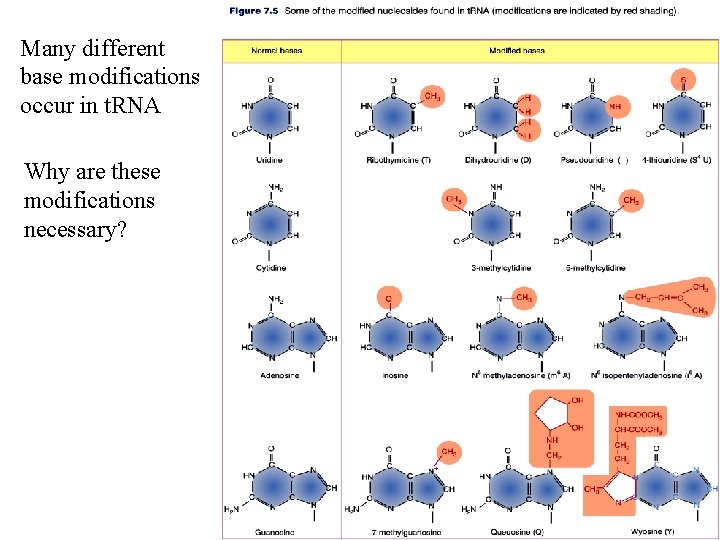
Many different base modifications occur in t. RNA Why are these modifications necessary?


Transfer RNA (t. RNA) and the genetic code Legitimate G-U bp Wobble bp


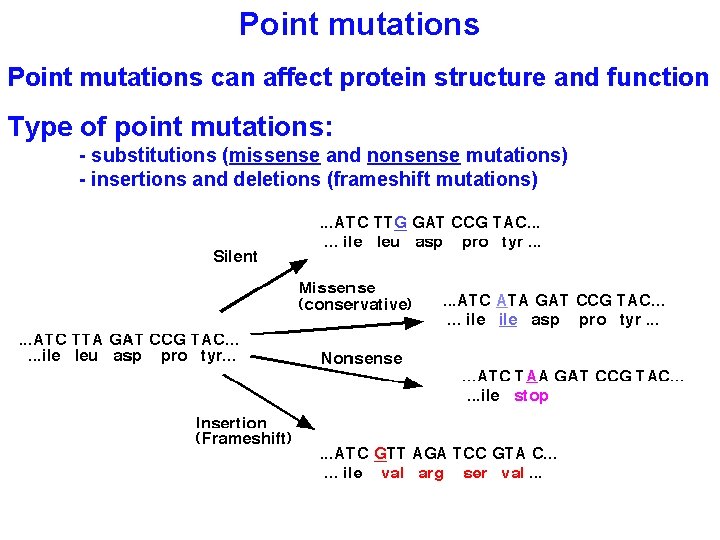
Point mutations can affect protein structure and function Type of point mutations: - substitutions (missense and nonsense mutations) - insertions and deletions (frameshift mutations)

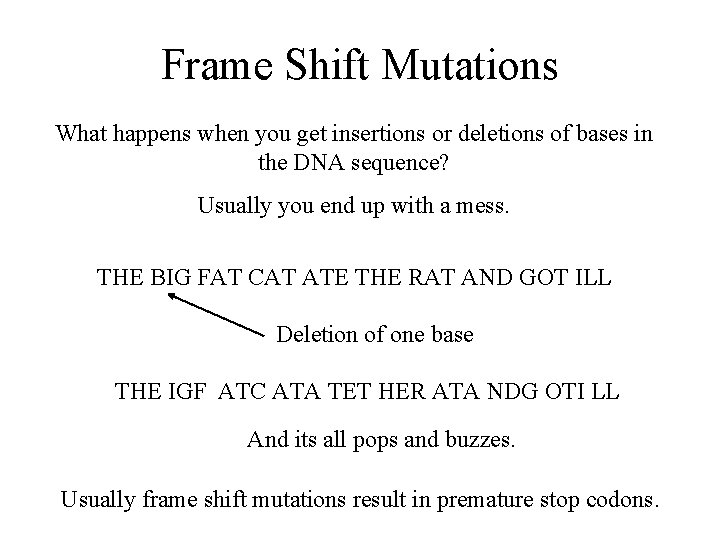
Frame Shift Mutations What happens when you get insertions or deletions of bases in the DNA sequence? Usually you end up with a mess. THE BIG FAT CAT ATE THE RAT AND GOT ILL Deletion of one base THE IGF ATC ATA TET HER ATA NDG OTI LL And its all pops and buzzes. Usually frame shift mutations result in premature stop codons.

Where can you get more information about the basic concepts embedded within the Central Dogma of Molecular Biology? Here is a great site, full of simple, clear, and animated (!) tutorials: http: //www. dnaftb. org/dnaftb/ Chapters 15 -28 in this series provides an excellent review of the first six lectures in Bioinformatics: http: //www. dnaftb. org/dnaftb/15/concept/

Ribosomes are large ribonucleoprotein (RNP) complexes They are complex affairs, composed of an array of RNA + proteins

