How cells use DNA part 2 TRANSLATION An




- Slides: 21

How cells use DNA, part 2: TRANSLATION

An overview: Most commonly, what comes to mind is the process by which we take ideas expressed in one language, & make them intelligible in another language. In the process of translation in a cell, the transcribed message of m. RNA is translated to a totally different ‘language’, that of protein. Often this means a change of script, from one we don’t understand to another we can read. DNA & RNA are ‘written’ in very similar chemicals, but protein is ‘written’ in an entirely different ‘script’: amino acids.

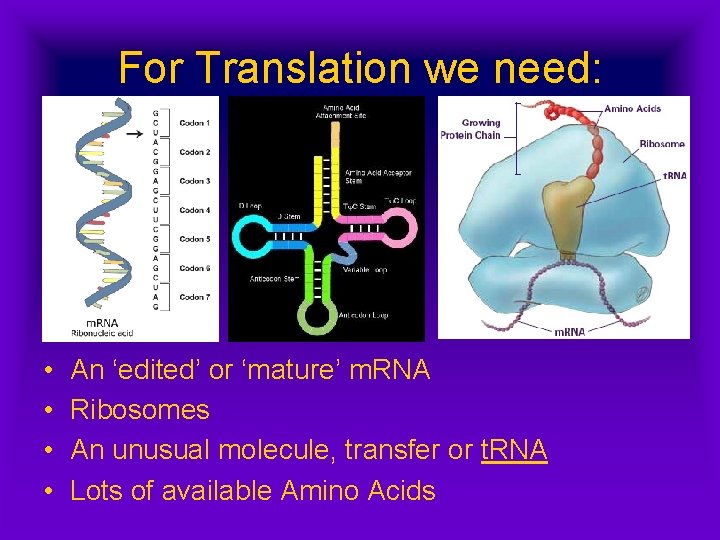
For Translation we need: • • An ‘edited’ or ‘mature’ m. RNA Ribosomes An unusual molecule, transfer or t. RNA Lots of available Amino Acids

The overall goal: • Use the DNA message that was copied out into m. RNA to produce a polypeptide or protein. • This is the second part of the CENTRAL DOGMA • It relies on the GENETIC CODE.

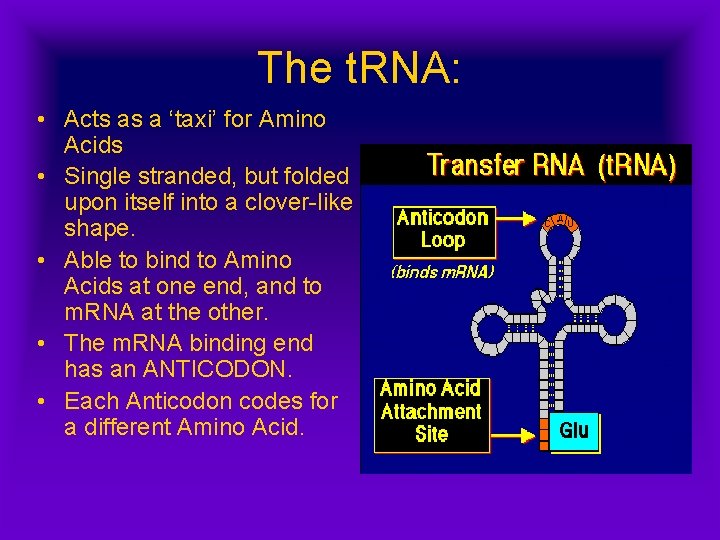
The t. RNA: • Acts as a ‘taxi’ for Amino Acids • Single stranded, but folded upon itself into a clover-like shape. • Able to bind to Amino Acids at one end, and to m. RNA at the other. • The m. RNA binding end has an ANTICODON. • Each Anticodon codes for a different Amino Acid.

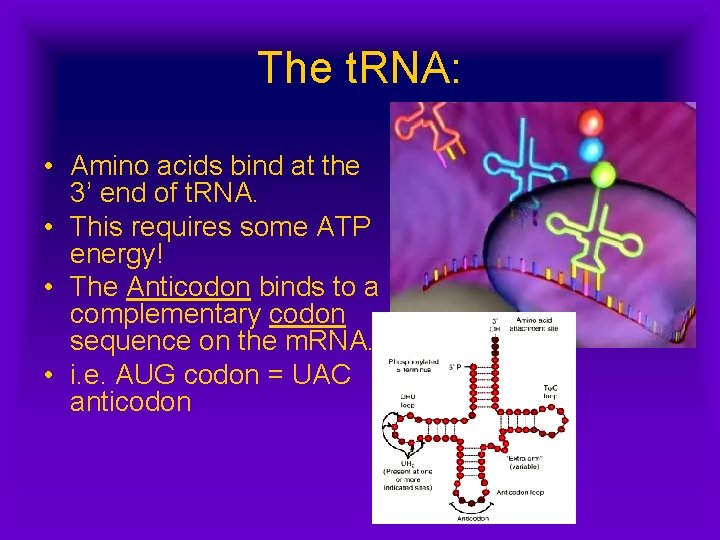
The t. RNA: • Amino acids bind at the 3’ end of t. RNA. • This requires some ATP energy! • The Anticodon binds to a complementary codon sequence on the m. RNA. • i. e. AUG codon = UAC anticodon

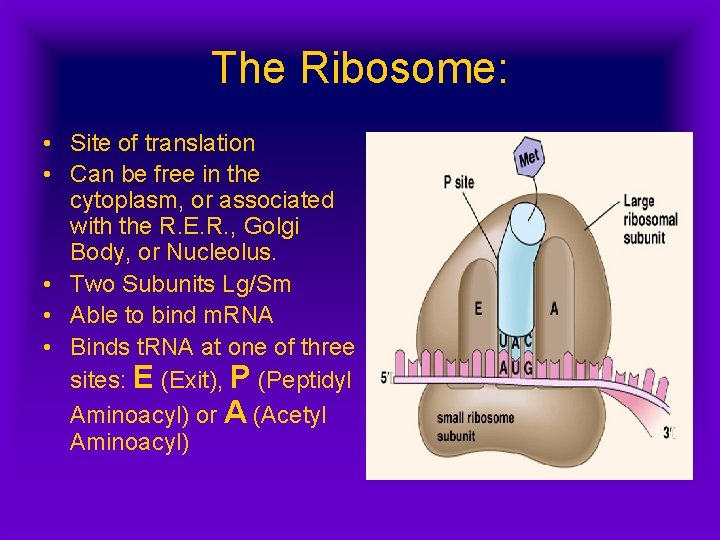
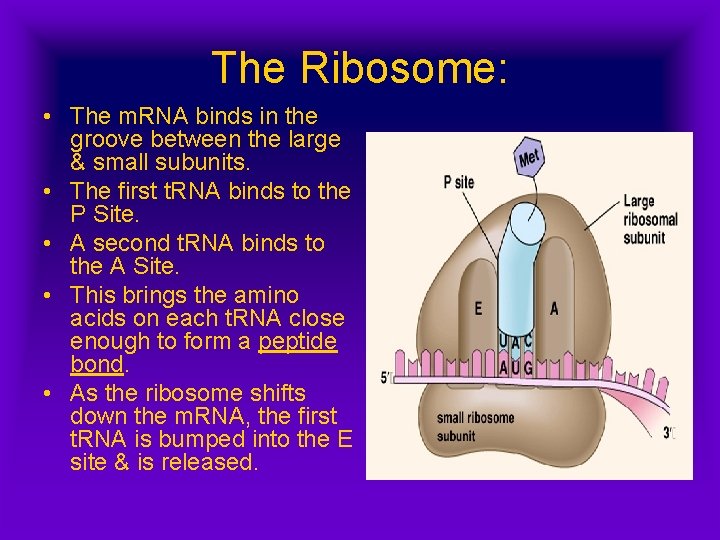
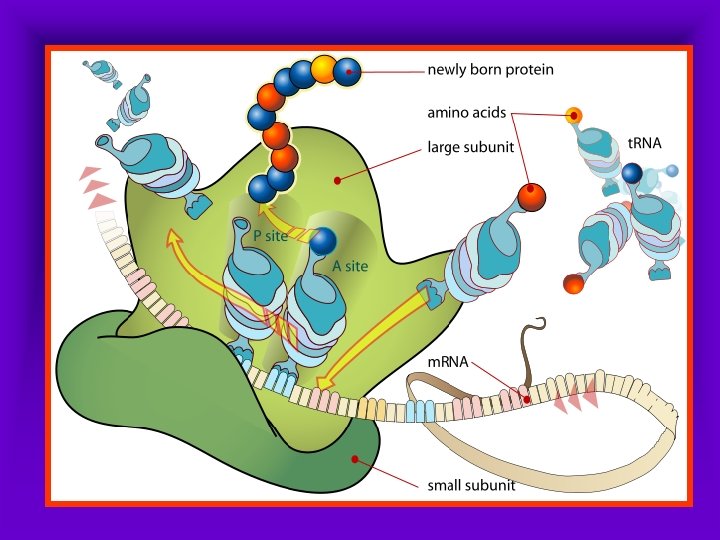
The Ribosome: • Site of translation • Can be free in the cytoplasm, or associated with the R. E. R. , Golgi Body, or Nucleolus. • Two Subunits Lg/Sm • Able to bind m. RNA • Binds t. RNA at one of three sites: E (Exit), P (Peptidyl Aminoacyl) or A (Acetyl Aminoacyl)

The Ribosome: • The m. RNA binds in the groove between the large & small subunits. • The first t. RNA binds to the P Site. • A second t. RNA binds to the A Site. • This brings the amino acids on each t. RNA close enough to form a peptide bond. • As the ribosome shifts down the m. RNA, the first t. RNA is bumped into the E site & is released.

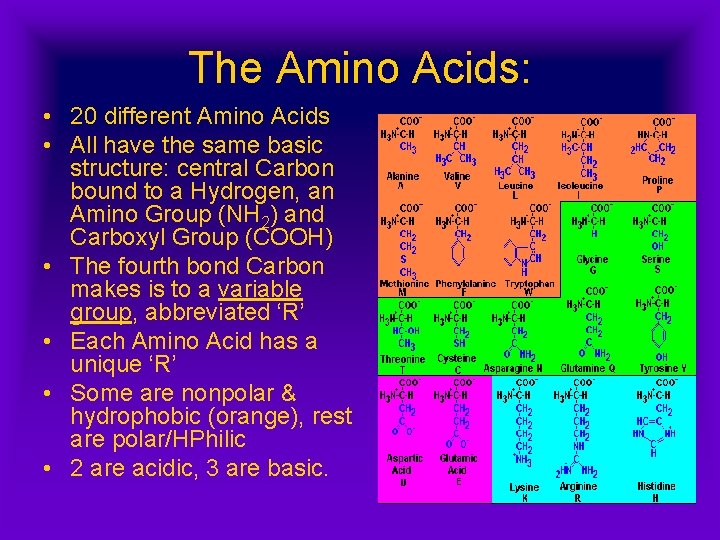
The Amino Acids: • 20 different Amino Acids • All have the same basic structure: central Carbon bound to a Hydrogen, an Amino Group (NH 2) and Carboxyl Group (COOH) • The fourth bond Carbon makes is to a variable group, abbreviated ‘R’ • Each Amino Acid has a unique ‘R’ • Some are nonpolar & hydrophobic (orange), rest are polar/HPhilic • 2 are acidic, 3 are basic.

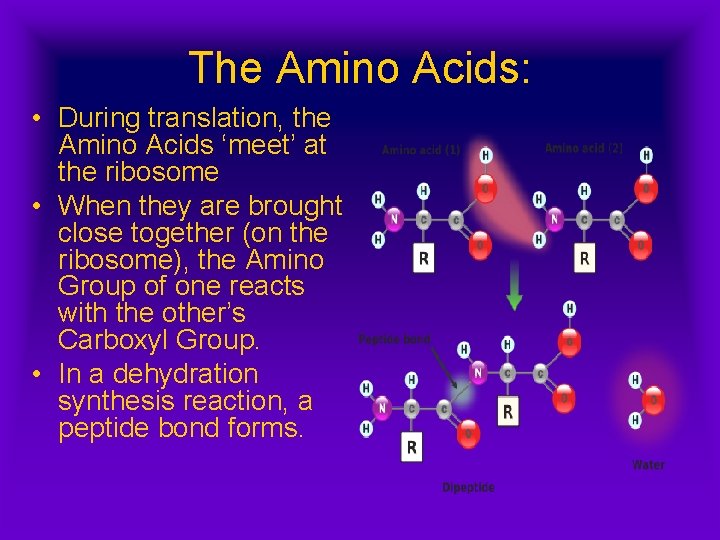
The Amino Acids: • During translation, the Amino Acids ‘meet’ at the ribosome • When they are brought close together (on the ribosome), the Amino Group of one reacts with the other’s Carboxyl Group. • In a dehydration synthesis reaction, a peptide bond forms.

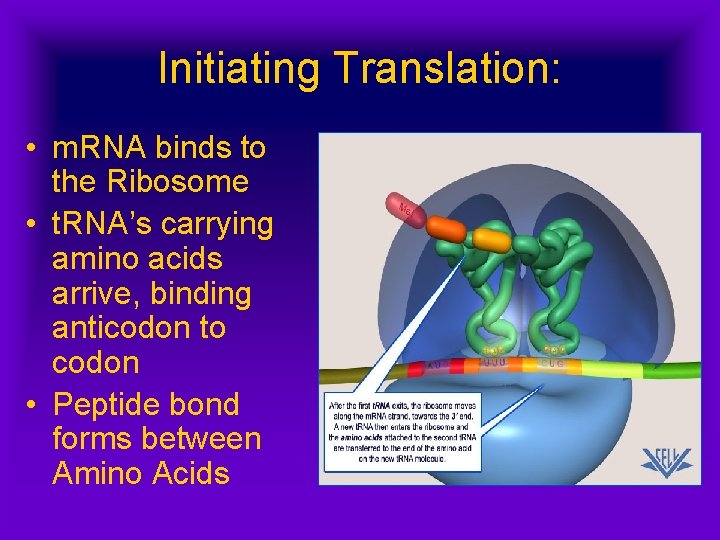
Initiating Translation: • m. RNA binds to the Ribosome • t. RNA’s carrying amino acids arrive, binding anticodon to codon • Peptide bond forms between Amino Acids

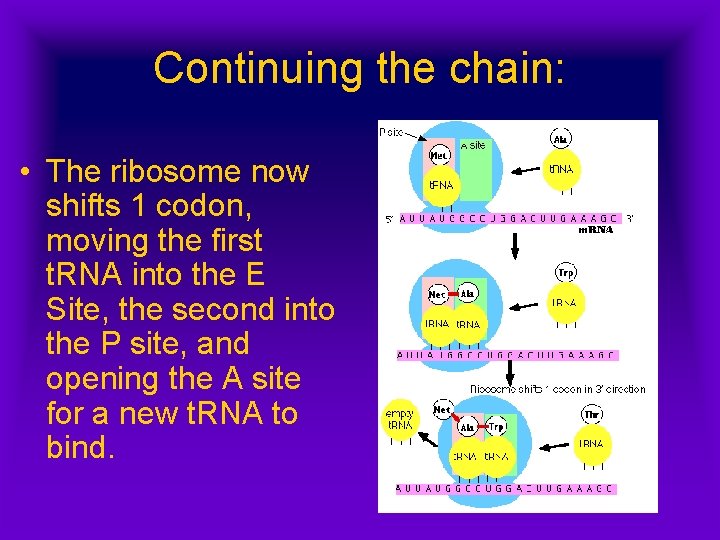
Continuing the chain: • The ribosome now shifts 1 codon, moving the first t. RNA into the E Site, the second into the P site, and opening the A site for a new t. RNA to bind.

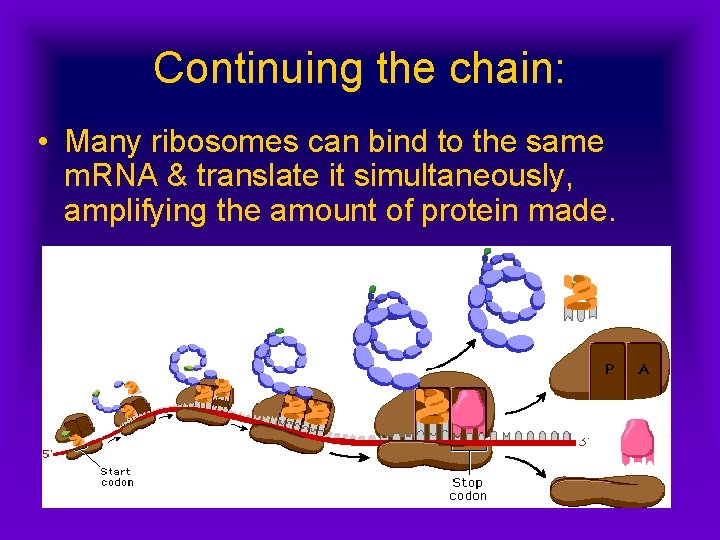
Continuing the chain: • Many ribosomes can bind to the same m. RNA & translate it simultaneously, amplifying the amount of protein made.

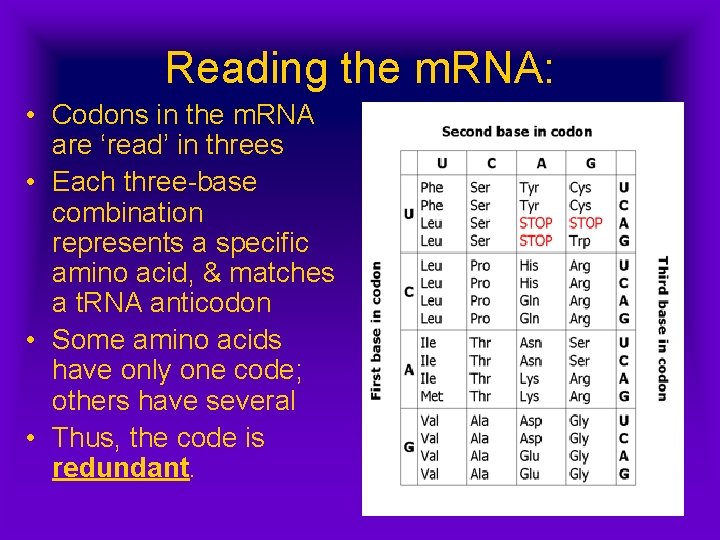
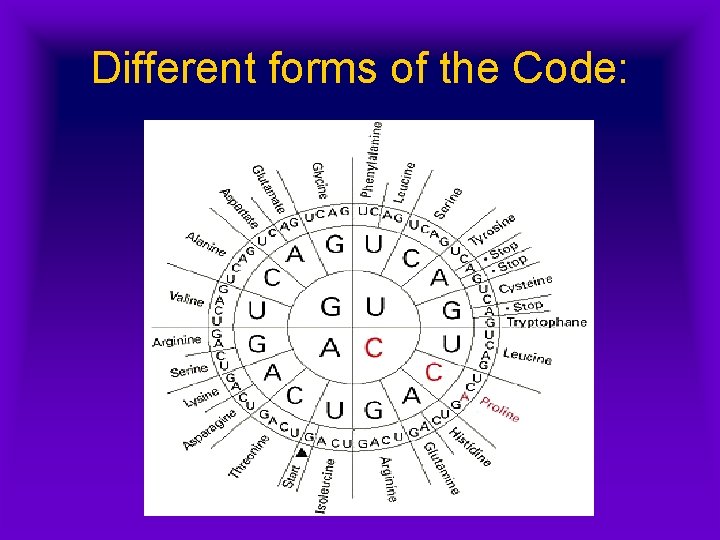
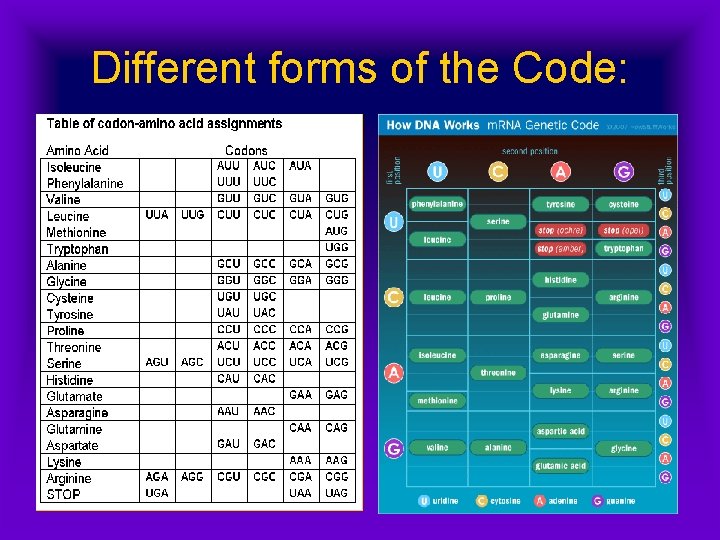
Reading the m. RNA: • Codons in the m. RNA are ‘read’ in threes • Each three-base combination represents a specific amino acid, & matches a t. RNA anticodon • Some amino acids have only one code; others have several • Thus, the code is redundant.

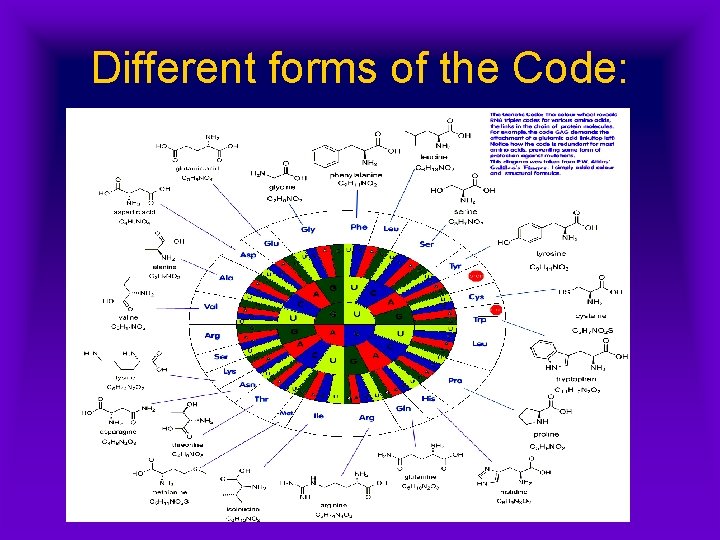
Different forms of the Code:

Different forms of the Code:

Different forms of the Code:

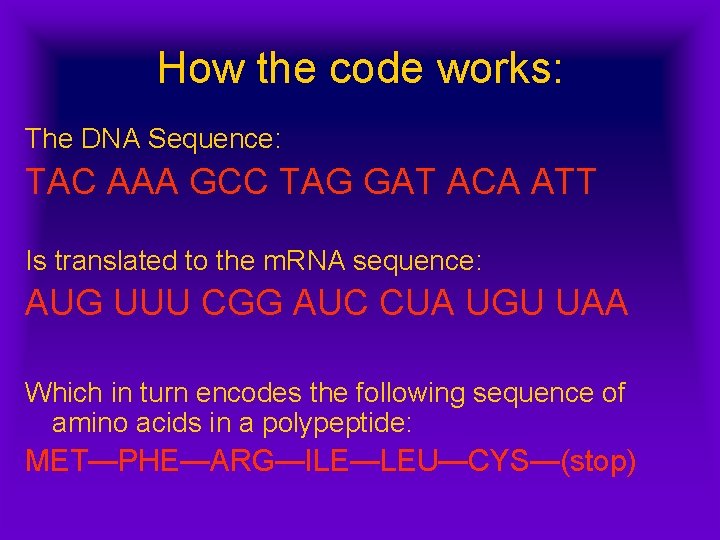
How the code works: The DNA Sequence: TAC AAA GCC TAG GAT ACA ATT Is translated to the m. RNA sequence: AUG UUU CGG AUC CUA UGU UAA Which in turn encodes the following sequence of amino acids in a polypeptide: MET—PHE—ARG—ILE—LEU—CYS—(stop)

Wrapping things up: • There are three m. RNA codons that signal the end of a protein • They are called STOP CODONS: UAA, UAG, UGA • *in DNA, these are ATT, ATC, & ACT. • When it reaches a stop codon, the ribosome releases the m. RNA, & translation ends.


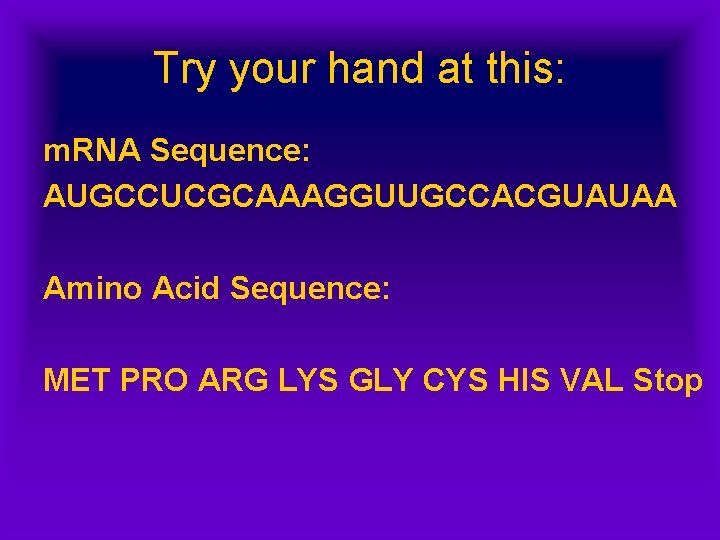
Try your hand at this: m. RNA Sequence: AUGCCUCGCAAAGGUUGCCACGUAUAA Amino Acid Sequence: MET PRO ARG LYS GLY CYS HIS VAL Stop