Homology If twp proteins are homologous they have

- Slides: 4

Homology If twp proteins are homologous, they have a common fold and a common ancestor If two proteins have >25% identity across their entire length, they are likely to be Homologs. However, sometimes true homologs have quite low sequence identity! Orthologs Paralogs Alignments Homologous (and equivalent) proteins from different species. Arise from speciation. Homologous (and equivalent) proteins found in same species. Divergence of sequences NOT from speciation. How to score? Minimum # of mutations? , Physicochemical properties (as perceived by us)? , Or learn from nature? Scoring schemes PAM, BLOSUM Gap penalties, low sequence complexity filtering

E values What it means in words E = Kmne -λS Alignment algorithms Why use local alignment algorithm? BLAST (Basic Local Alignment Search Tool) FASTA (Fast Alignment) Smith-Waterman Needleman-Wunsch

IF A is related to B and B is related to C, then A is related to C (use orthologs, paralogs, known homologs PSI BLAST 3 D-PSSM 1 normal BLAST run then subsequent iterations modify the scoring matrix – starts “rewarding” conservation at key positions in the alignment Database of structures grouped into fold families (homologs) Careful 1 D PSSM is created for each fold family. The 1 D PSSM is augmented by info from structural alignment of all family members(3 D-PSSM). Also, entire library is used to assign solvation potentials (how likely is a glutamate to be only 5% exposed? 10% exposed. Etc. ) Query sequence Get 1 D-PSSM from nr database. Do 2 ndry structure prediction. Now compare the query sequence to each library entry in 3 ways: Use the library Entry’s 1 D-PSSM(augmented), see how the query compares to the 3 D-PSSM of The library entry. See how the library entry compares to the 1 D-PSSM for the query.

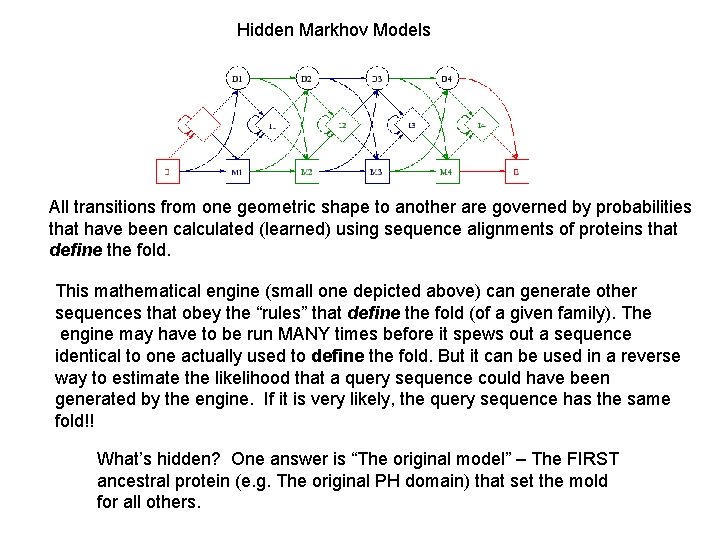
Hidden Markhov Models All transitions from one geometric shape to another are governed by probabilities that have been calculated (learned) using sequence alignments of proteins that define the fold. This mathematical engine (small one depicted above) can generate other sequences that obey the “rules” that define the fold (of a given family). The engine may have to be run MANY times before it spews out a sequence identical to one actually used to define the fold. But it can be used in a reverse way to estimate the likelihood that a query sequence could have been generated by the engine. If it is very likely, the query sequence has the same fold!! What’s hidden? One answer is “The original model” – The FIRST ancestral protein (e. g. The original PH domain) that set the mold for all others.