Homework Wednesday 92706 Due on Wednesday 10406 1











- Slides: 19

Homework (Wednesday, 9/27/06, Due on Wednesday, 10/4/06) 1. Show the molecular orbitals of P 15. 2. Calculate the exact molecular weights of each of the four standard nucleotides (A, T, G, and C) in DNA molecules? 3. p. BR 322 is a circular double-stranded plasmid of 4363 nucleotides. Figure out 1) how many picomoles are 1 microgram of the plasmid? 2) How many micrograms are 1 pmol of the plasmid? 3) How much are the weights of two single-stranded circular p. BBR 322, respectively? 4. Draw diagrams to show all possible base-pairings of adenine and thymine.

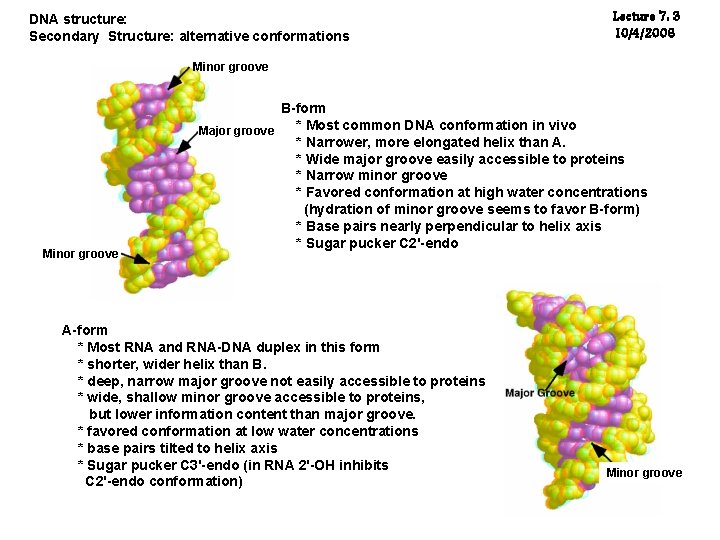
Lecture 7: 1 10/4/2006 DNA structure: Secondary Structure: alternative conformations Minor groove The minor groove is generated by the smaller angular distance between sugars. Major groove major groove: The larger of the two grooves that spiral around the surface of the B-form of DNA.

DNA structure: Secondary Structure: alternative conformations Lecture 7: 2 10/4/2006

DNA structure: Secondary Structure: alternative conformations Lecture 7: 3 10/4/2006 Minor groove B-form * Most common DNA conformation in vivo Major groove * Narrower, more elongated helix than A. * Wide major groove easily accessible to proteins * Narrow minor groove * Favored conformation at high water concentrations (hydration of minor groove seems to favor B-form) * Base pairs nearly perpendicular to helix axis * Sugar pucker C 2'-endo A-form * Most RNA and RNA-DNA duplex in this form * shorter, wider helix than B. * deep, narrow major groove not easily accessible to proteins * wide, shallow minor groove accessible to proteins, but lower information content than major groove. * favored conformation at low water concentrations * base pairs tilted to helix axis * Sugar pucker C 3'-endo (in RNA 2'-OH inhibits C 2'-endo conformation) Minor groove

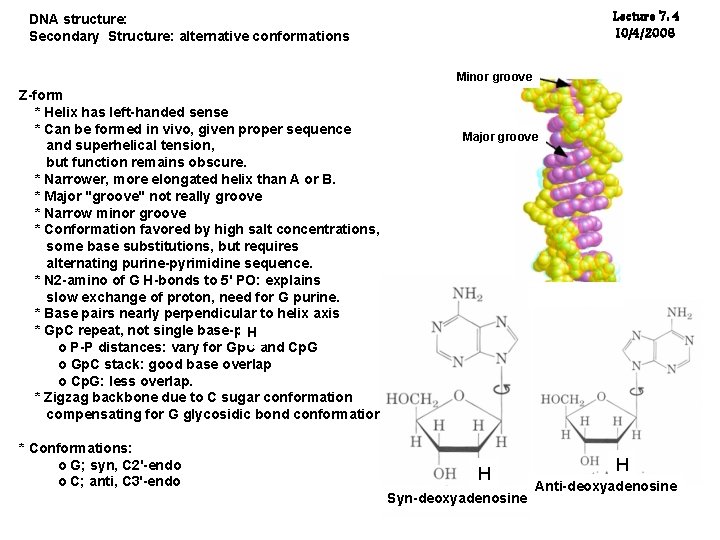
Lecture 7: 4 10/4/2006 DNA structure: Secondary Structure: alternative conformations Minor groove Z-form * Helix has left-handed sense * Can be formed in vivo, given proper sequence and superhelical tension, but function remains obscure. * Narrower, more elongated helix than A or B. * Major "groove" not really groove * Narrow minor groove * Conformation favored by high salt concentrations, some base substitutions, but requires alternating purine-pyrimidine sequence. * N 2 -amino of G H-bonds to 5' PO: explains slow exchange of proton, need for G purine. * Base pairs nearly perpendicular to helix axis * Gp. C repeat, not single base-pair H o P-P distances: vary for Gp. C and Cp. G o Gp. C stack: good base overlap o Cp. G: less overlap. * Zigzag backbone due to C sugar conformation compensating for G glycosidic bond conformation * Conformations: o G; syn, C 2'-endo o C; anti, C 3'-endo Major groove H H Syn-deoxyadenosine H H Anti-deoxyadenosine

DNA structure: Secondary Structure: alternative conformations Lecture 7: 5 10/4/2006 In Z-DNA and Z-RNA the (desoxy)ribo-phophate backbone forms a left-handed, zigzag helix where the glycosidic bonds between base and sugar alternate between the syn- and anti-conformation and the purines are tilted to the outside. In contrast, in B-DNA and A-RNA the backbone forms a smooth right-handed helix and all nucleotides are in the anti-conformation facing towards the inside of the helix. At the boundary between B-DNA and Z-DNA, a B-Z junction is formed in which a base pair is broken and the bases are extruded on each side. Cellular functions of Z-DNA: 1. Z-DNA can either enhance or repress promoter activity. 2. Z-DNA and Z-RNA can be bound by Z-DNA binding proteins (ZBPs), modulating immune functions. …………………………………………. . .

Lecture 7: 6 10/4/2006 DNA structure: Secondary Structure: visualizing DNA by X-Ray crystallography An average chemical bond has an average bond of 1. 5 angstroms. X-ray is photons with a wavelength ranging from 0. 05 to 100 angstroms. Filaments Detector Electrons A crystal Diffracted X-Ray Metal target (Cu) Primary X-ray (Cu. Kα) or from synchrotrons Grow crystals by hanging drop setup

DNA Secondary structure: Secondary Structure: visualizing DNA by X-Ray crystallography Lecture 7: 7 10/4/2006 A synchrotron is a particular type of cyclic particle accelerator in which the magnetic field (to turn the particles so they circulate) and the electric field (to accelerate the particles) are carefully synchronized with the traveling particle beam. Currently, the highest energy synchrotron in the world is the Tevatron, at the Fermi National Accelerator Laboratory, in the United States. It accelerates protons and antiprotons to slightly less than 1 Te. V of kinetic energy and collides them together. The Large Hadron Collider (LHC), which is being built at the European Laboratory for High Energy Physics (CERN), will have roughly seven times this energy, and is scheduled to turn on in 2007. It is being built in the 27 km tunnel which formerly housed the Large Electron Positron (LEP) collider, so it will maintain the claim as the largest scientific device ever built. The LHC will also accelerate heavy ions (such as Lead) up to an energy of 1150 Te. V. The largest device of this type seriously proposed was the Superconducting Super Collider (SSC), ……………………………… Structure biologists use synchrotron radiation (synchrotron X-rays) to illuminate the crystals of biological samples. Advantages: 1) Small crystals; 2) Short exposures

DNA Secondary structure: Secondary Structure: visualizing DNA by X-Ray crystallography Lecture 7: 8 10/4/2006 In 1979, Rich and co-workers at MIT accidentally grew a crystal of ZDNA[1]. This was the first crystal structure of any form of DNA. After 26 years of attempts, Rich et al. finally crystallized the junction box of B- and Z-DNA. Their results were published in an October 2005 Nature journal[2]. Whenever Z-DNA forms, there must be two junction boxes that allow the flip back to the canonical B-form of DNA. [1]. Wang AHJ, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, Van der Marel G, and Rich A (1979). Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature (London) 282: 680 -686. [2]. Ha SC, Lowenhaupt K, Rich A, Kim YG, and Kim KK (2005). Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature 437: 1183 -1186.

Lecture 7: 9 10/6/2006 DNA structure: Secondary Structure: comparisons of alternative forms Properties of different helical forms Geometry attribute Helix sense A-form B-form Z-form right-handed left-handed Repeating unit 1 bp 2 bp 33. 6° 35. 9° (± 4. 2°) 60°/2 Mean bp/turn 10. 7 10. 0 (± 1. 2) 12 Inclination of bp to axis +19° -1. 2° (± 4. 1°) -9° Rise/bp along axis 0. 23 nm 0. 332 nm (± 0. 019 nm) 0. 38 nm Pitch/turn of helix 2. 46 nm 3. 32 nm (± 0. 19 nm) 4. 56 nm +18° +16° ( ± 7°) 0° Rotation/bp Mean propeller twist Glycosyl angle anti C: anti, G: syn Sugar pucker C 3'-endo C 2'-endo C: C 2'-endo, G: C 2'-exo Diameter 2. 55 nm 2. 37 nm 1. 84 nm

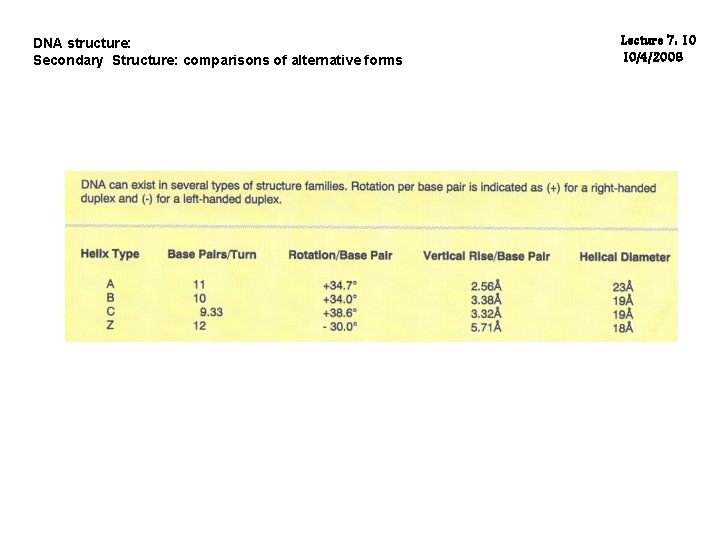
DNA structure: Secondary Structure: comparisons of alternative forms Lecture 7: 10 10/4/2006

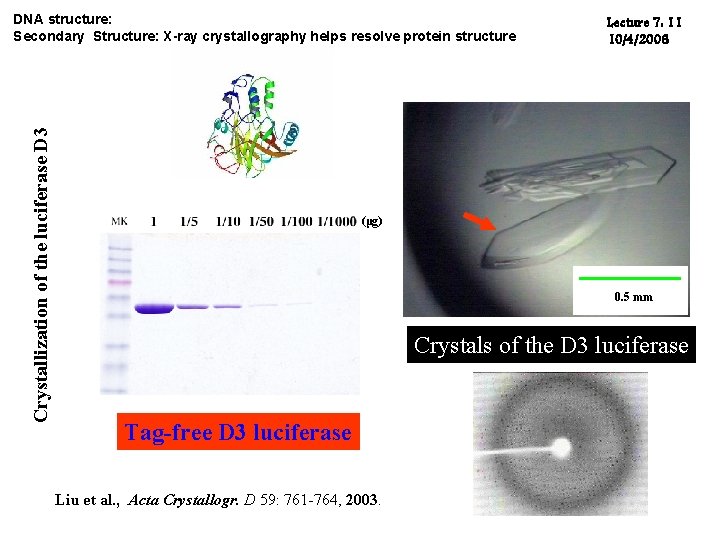
Crystallization of the luciferase D 3 DNA structure: Secondary Structure: X-ray crystallography helps resolve protein structure Lecture 7: 11 10/4/2006 (μg) 0. 5 mm Crystals of the D 3 luciferase Tag-free D 3 luciferase Liu et al. , Acta Crystallogr. D 59: 761 -764, 2003.

DNA structure: Secondary Structure: NMR helps resolve DNA structures Lecture 7: 12 10/4/2006 Spectroscopy is the study of the interaction of electromagnetic radiation with matter. Nuclear magnetic resonance spectroscopy is the use of the NMR phenomenon to study physical, chemical, and biological properties of matter. Spin What is spin? Spin is a fundamental property of nature like electrical charge or mass. Spin comes in multiples of 1/2 and can be + or -. Protons, electrons, and neutrons possess spin. Individual unpaired electrons, protons, and neutrons each possesses a spin of 1/2. In the deuterium atom ( 2 H ), with one unpaired electron, one unpaired proton, and one unpaired neutron, the total electronic spin = 1/2 and the total nuclear spin = 1. When placed in a magnetic field of strength B, a particle with a net spin can absorb a photon, of frequency. The frequency ν depends on the gyromagnetic ratio, γ of the particle. ν = , γ B For hydrogen, = 42. 58 MHz / T. Nuclei with Spin The shell model for the nucleus tells us that nucleons, just like electrons, fill orbitals. When the number of protons or neutrons equals 2, 8, 20, 28, 50, 82, and 126, orbitals are filled. Because nucleons have spin, just like electrons do, their spin can pair up when the orbitals are being filled and cancel out. Energy Levels To understand how particles with spin behave in a magnetic field, consider a proton. This proton has the property called spin. Think of the spin of this proton as a magnetic moment vector, causing the proton to behave like a tiny magnet with a north and south pole

DNA structure: Secondary Structure: NMR helps resolve DNA structures 1. A spinning charge generates a magnetic field, as shown by the animation on the right. The resulting spin-magnet has a magnetic moment (μ) proportional to the spin. 2. In the presence of an external magnetic field (B 0), two spin states exist, +1/2 and 1/2. The magnetic moment of the lower energy +1/2 state is aligned with the external field, but that of the higher energy -1/2 spin state is opposed to the external field. Note that the arrow representing the external field points North 3. The difference in energy between the two spin states is dependent on the external magnetic field strength, and is always very small. The following diagram illustrates that the two spin states have the same energy when the external field is zero, but diverge as the field increases. At a field equal to Bx a formula for the energy difference is given (remember I = 1/2 and μ is the magnetic moment of the nucleus in the field). 4. For spin 1/2 nuclei the energy difference between the two spin states at a given magnetic field strength will be proportional to their magnetic moments. Lecture 7: 13 10/4/2006

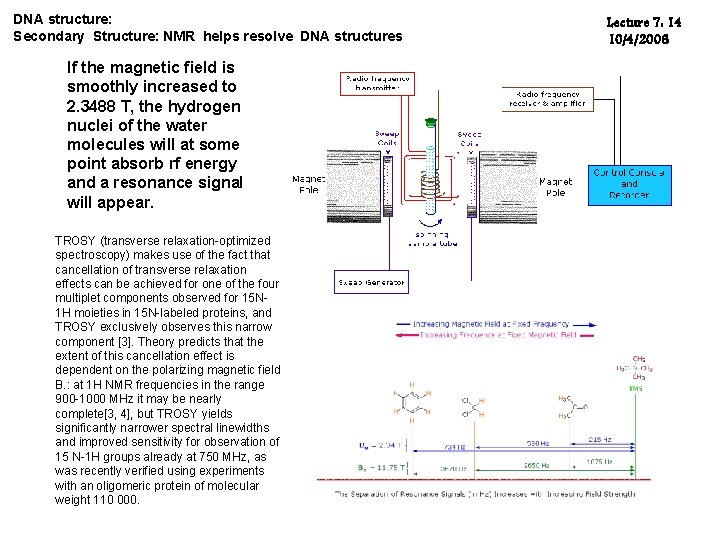
DNA structure: Secondary Structure: NMR helps resolve DNA structures If the magnetic field is smoothly increased to 2. 3488 T, the hydrogen nuclei of the water molecules will at some point absorb rf energy and a resonance signal will appear. TROSY (transverse relaxation-optimized spectroscopy) makes use of the fact that cancellation of transverse relaxation effects can be achieved for one of the four multiplet components observed for 15 N 1 H moieties in 15 N-labeled proteins, and TROSY exclusively observes this narrow component [3]. Theory predicts that the extent of this cancellation effect is dependent on the polarizing magnetic field B. : at 1 H NMR frequencies in the range 900 -1000 MHz it may be nearly complete[3, 4], but TROSY yields significantly narrower spectral linewidths and improved sensitivity for observation of 15 N-1 H groups already at 750 MHz, as was recently verified using experiments with an oligomeric protein of molecular weight 110 000. Lecture 7: 14 10/4/2006

DNA structure: Secondary Structure: Base Stacking The electrons are actually shared in such a pattern, so that the density between all the atoms is close to 1. 5 bonds, and the p orbitals overlap to form a system called a pi orbital. The simple benzene molecule, where all six atoms are equivalent, is pictured at the right. The electron density is uniformly distributed around the ring, and one component of the pi-electron system is the sum of the electron lobes seen here. The sum of all pi-orbitals is symmetric above and below the ring. Rings that stack share pi-electrons, resulting in a favorable configuration. Stacks of planer systems with multiple conjugated rings are favored even more, thus the purines, particularly guanine, have high stacking energies. Base Stacking: 1. When the bases are stacked, the charges contribute to the stacking energy. 2. the electrostatic field of one base induce attractive, asymmetrical electronic distributions in the electron distribution of the other base. 3. Spontaneous fluctuations in charge occur in each base and induce dipoles in the other. Lecture 7: 15 10/4/2006

DNA structure: Secondary Structure: Triple helical DNA Lecture 7: 16 10/4/2006 Coordination of transition metal cations to purine nucleotides in the third strand may cause polarization of the base charges, eventually leading to stronger hydrogen bonds with the complementary purine nucleotides in one strand of the target DNA duplex, thereby strengthening the triplex structure. Another source of triplex stability is related to stretches of cationic amino acid residues in proteins.

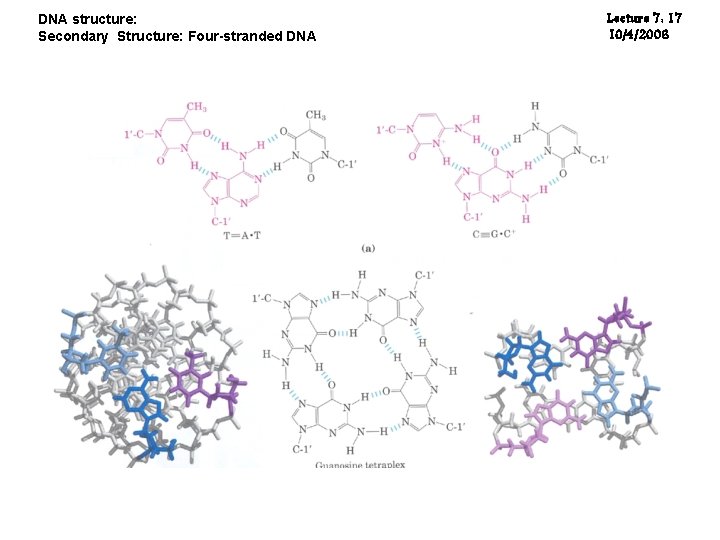
DNA structure: Secondary Structure: Four-stranded DNA Lecture 7: 17 10/4/2006

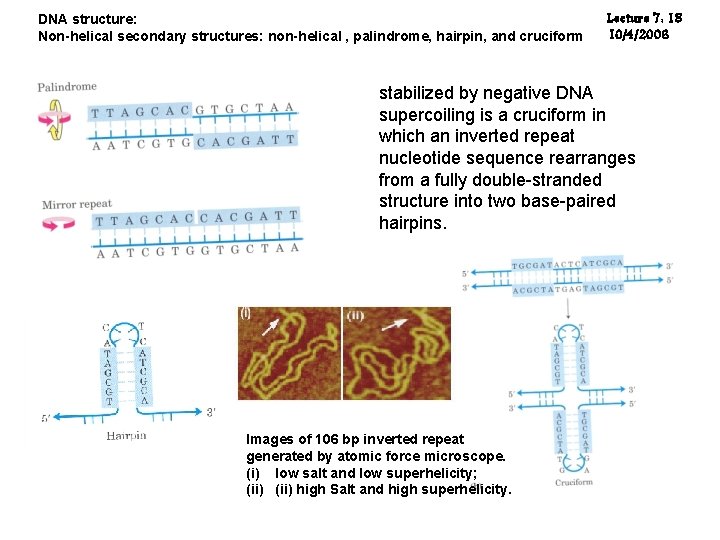
DNA structure: Non-helical secondary structures: non-helical , palindrome, hairpin, and cruciform Lecture 7: 18 10/4/2006 stabilized by negative DNA supercoiling is a cruciform in which an inverted repeat nucleotide sequence rearranges from a fully double-stranded structure into two base-paired hairpins. Images of 106 bp inverted repeat generated by atomic force microscope. (i) low salt and low superhelicity; (ii) high Salt and high superhelicity.
 Literal language examples
Literal language examples Parts of a poem
Parts of a poem Homework i love you poem
Homework i love you poem Homework oh homework i hate you you stink
Homework oh homework i hate you you stink Alitteration definition
Alitteration definition Homework oh homework i hate you you stink
Homework oh homework i hate you you stink Homework due today
Homework due today Homework due today
Homework due today Homework is due
Homework is due Folk culture and popular culture venn diagram
Folk culture and popular culture venn diagram Homework due today
Homework due today Astr
Astr Iron core
Iron core Homework is due on friday
Homework is due on friday Homework due tomorrow
Homework due tomorrow Homework due today
Homework due today Homework is due on friday
Homework is due on friday Quadrilatero con due angoli retti
Quadrilatero con due angoli retti Liberty chapter 20
Liberty chapter 20 Grande rhetra
Grande rhetra