HL 7 v 3 Clinical Genomics Overview The











- Slides: 52

HL 7 v 3 Clinical Genomics – Overview The HL 7 Clinical Genomics Work Group Prepared by Amnon Shabo (Shvo), Ph. D HL 7 Clinical Genomics WG Co-chair and Modeling Facilitator HL 7 Structured Documents WG CDA Co-editor CCD Implementation Guide Co-editor GTR Implementation Guide prime editor HL 7 RIMBAA WG, Co-chair

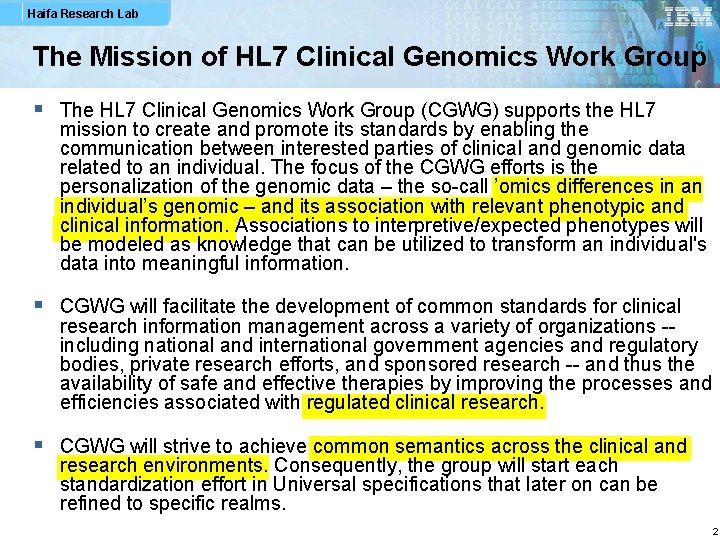
Haifa Research Lab The Mission of HL 7 Clinical Genomics Work Group § The HL 7 Clinical Genomics Work Group (CGWG) supports the HL 7 mission to create and promote its standards by enabling the communication between interested parties of clinical and genomic data related to an individual. The focus of the CGWG efforts is the personalization of the genomic data – the so-call ’omics differences in an individual’s genomic – and its association with relevant phenotypic and clinical information. Associations to interpretive/expected phenotypes will be modeled as knowledge that can be utilized to transform an individual's data into meaningful information. § CGWG will facilitate the development of common standards for clinical research information management across a variety of organizations -including national and international government agencies and regulatory bodies, private research efforts, and sponsored research -- and thus the availability of safe and effective therapies by improving the processes and efficiencies associated with regulated clinical research. § CGWG will strive to achieve common semantics across the clinical and research environments. Consequently, the group will start each standardization effort in Universal specifications that later on can be refined to specific realms. 2

Haifa Research Lab Overview of Activities Three Tracks: v 3: v 2: CDA: § Family History v 2 Implementation Guides § A CDA Implementation (Pedigree) Topic § Genetic Variations Topic § Gene Expression Topic § CMETs defined by the * The IG “Genetic Test Result Reporting to EHR” is modeled after the HL 7 Version 2. 5. 1 Implementation Guide: Orders And Observations; Interoperable Laboratory Result Reporting To EHR (US Realm), Release 1 Guide for Genetic Testing Reports Domain Common: § Domain Analysis Models for the various topics § A Domain Information Model (v 3) describing the common semantics Normative DSTU Informative § Semantic alignment among the various specs 3

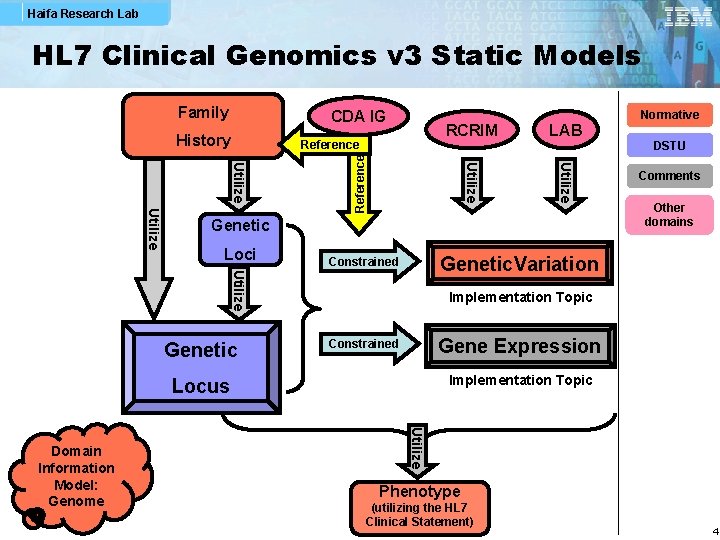
Haifa Research Lab HL 7 Clinical Genomics v 3 Static Models Family Utilize Genetic Loci Normative DSTU Comments Other domains Genetic. Variation Constrained Utilize Genetic Implementation Topic Gene Expression Constrained Implementation Topic Locus Utilize Domain Information Model: Genome LAB Reference RCRIM Utilize History Utilize CDA IG Phenotype (utilizing the HL 7 Clinical Statement) 4

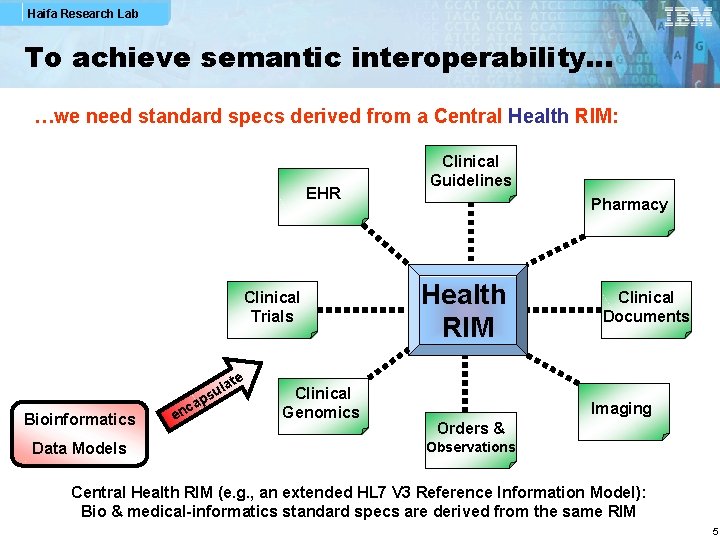
Haifa Research Lab To achieve semantic interoperability… …we need standard specs derived from a Central Health RIM: EHR Clinical Trials Bioinformatics Data Models c en te a l su ap Clinical Genomics Clinical Guidelines Pharmacy Health RIM Clinical Documents Imaging Orders & Observations Central Health RIM (e. g. , an extended HL 7 V 3 Reference Information Model): Bio & medical-informatics standard specs are derived from the same RIM 5

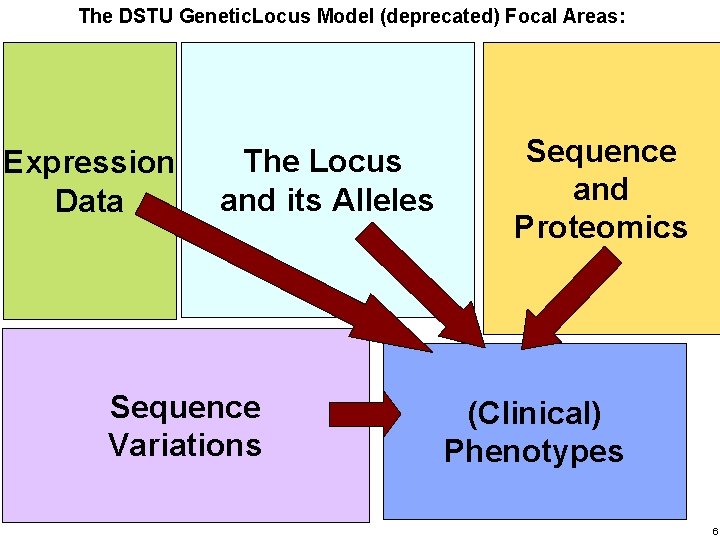
The DSTU Genetic. Locus Model (deprecated) Focal Areas: Haifa Research Lab Expression Data The Locus and its Alleles Sequence Variations Sequence and Proteomics (Clinical) Phenotypes 6

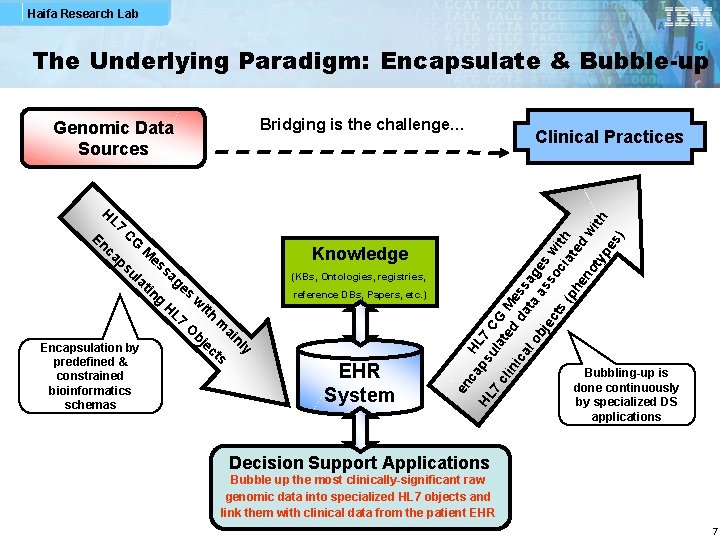
Haifa Research Lab The Underlying Paradigm: Encapsulate & Bubble-up Bridging is the challenge… predefined & constrained bioinformatics schemas Knowledge (KBs, Ontologies, registries, reference DBs, Papers, etc. ) EHR System ap HL su 7 C 7 la G cl te M in d ic da ess al ta ag ob as es je so w ct s ci ith (p at e he no d w ith ty pe s) En CG ca M es ps sa ul ge at in s g w H L 7 ith m O ai bj nl e y Encapsulation by ct s HL H L 7 Clinical Practices en c Genomic Data Sources Bubbling-up is done continuously by specialized DS applications Decision Support Applications Bubble up the most clinically-significant raw genomic data into specialized HL 7 objects and link them with clinical data from the patient EHR 7

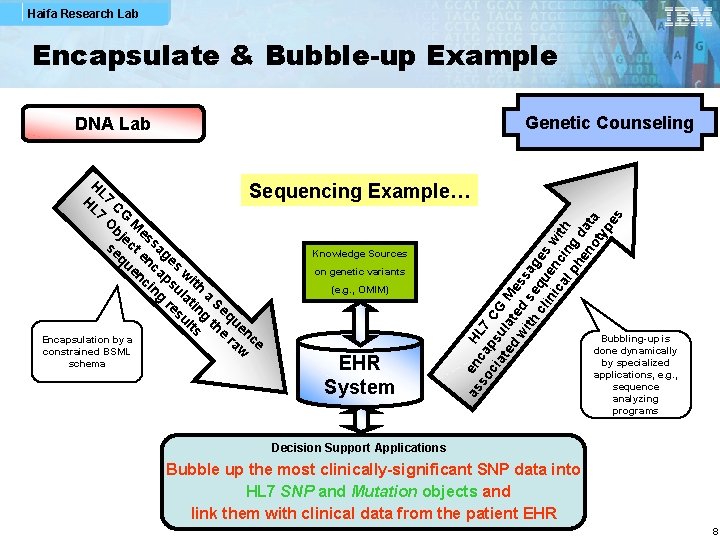
Haifa Research Lab Encapsulate & Bubble-up Example Genetic Counseling DNA Lab H schema EHR System as en HL so ca 7 ci ps CG at ul ed at M wi ed ess th se ag cl qu es in en w ic al cin ith ph g en da ot ta yp es L Sequencing Example… H 7 C L 7 G O M bj e e s se ct sag Knowledge Sources qu en es en ca w on genetic variants ci ps ith ng ul (e. g. , OMIM) a re atin Se su g q lts th ue e nc ra e Encapsulation by a w constrained BSML Bubbling-up is done dynamically by specialized applications, e. g. , sequence analyzing programs Decision Support Applications Bubble up the most clinically-significant SNP data into HL 7 SNP and Mutation objects and link them with clinical data from the patient EHR 8

Haifa Research Lab Omics in the LS DAM - Molecular Biology 9

Haifa Research Lab Omics in the LS DAM - Experiment 10

Haifa Research Lab Omics in the LS DAM - Specimen 11

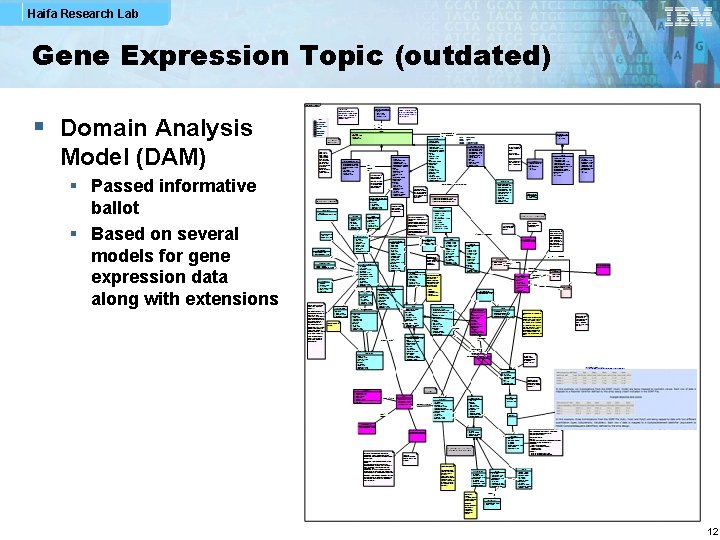
Haifa Research Lab Gene Expression Topic (outdated) § Domain Analysis Model (DAM) § Passed informative ballot § Based on several models for gene expression data along with extensions 12

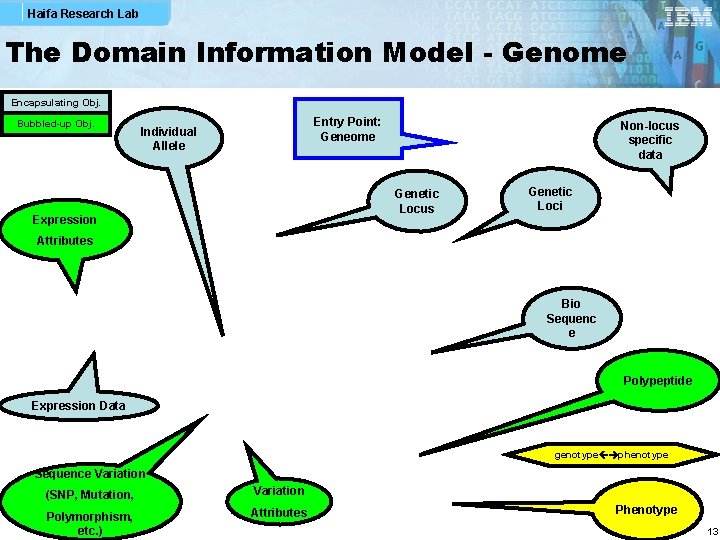
Haifa Research Lab The Domain Information Model - Genome Encapsulating Obj. Bubbled-up Obj. Entry Point: Geneome Individual Allele Non-locus specific data Genetic Locus Expression Genetic Loci Attributes Bio Sequenc e Polypeptide Expression Data genotype phenotype Sequence Variation (SNP, Mutation, Variation Polymorphism, etc. ) Attributes Phenotype 13

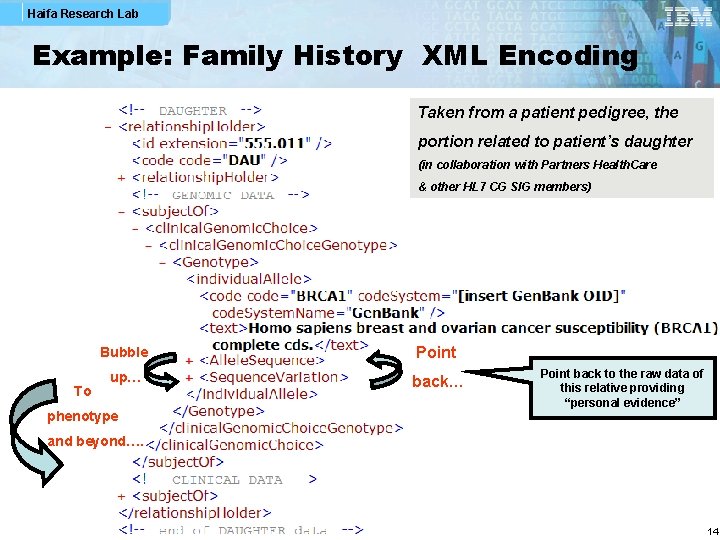
Haifa Research Lab Example: Family History XML Encoding Taken from a patient pedigree, the portion related to patient’s daughter (in collaboration with Partners Health. Care & other HL 7 CG SIG members) To Bubble Point up… back… phenotype Point back to the raw data of this relative providing “personal evidence” and beyond…. 14

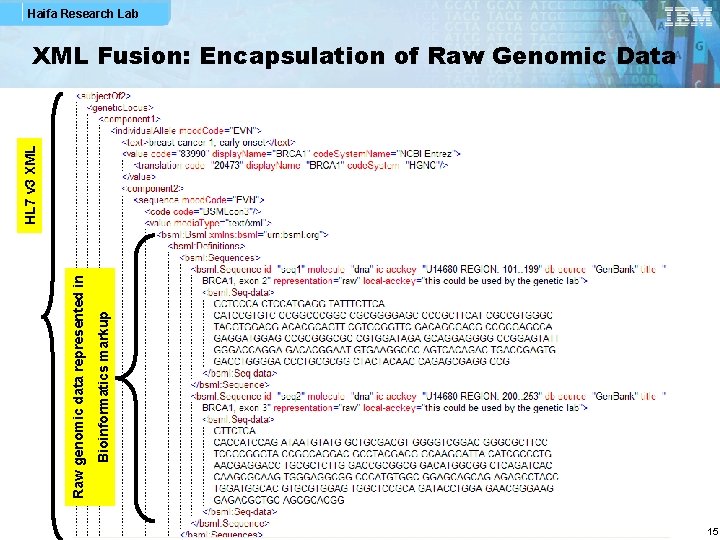
Haifa Research Lab Bioinformatics markup Raw genomic data represented in HL 7 v 3 XML Fusion: Encapsulation of Raw Genomic Data 15

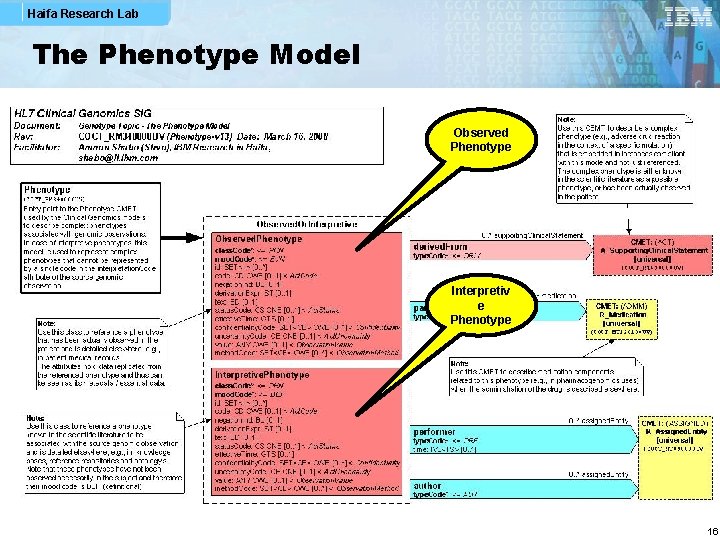
Haifa Research Lab The Phenotype Model Observed Phenotype Interpretiv e Phenotype 16

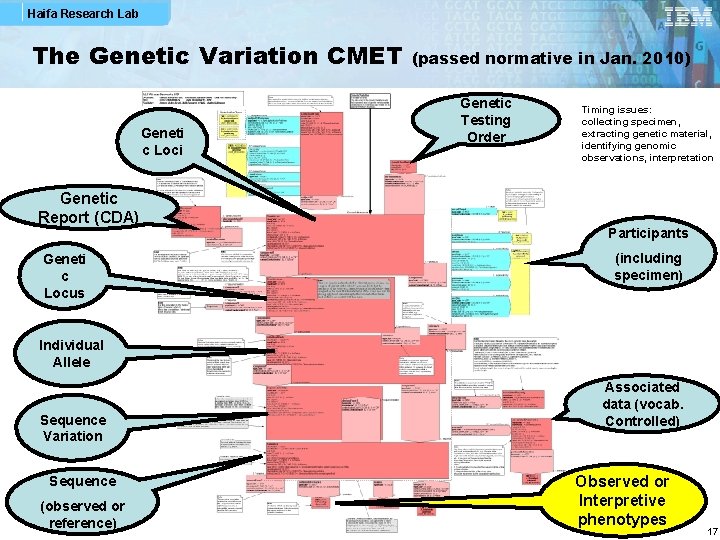
Haifa Research Lab The Genetic Variation CMET Geneti c Loci Genetic Report (CDA) Geneti c Locus (passed normative in Jan. 2010) Genetic Testing Order Timing issues: collecting specimen, extracting genetic material, identifying genomic observations, interpretation Participants (including specimen) Individual Allele Sequence Variation Sequence (observed or reference) Associated data (vocab. Controlled) Observed or Interpretive phenotypes 17

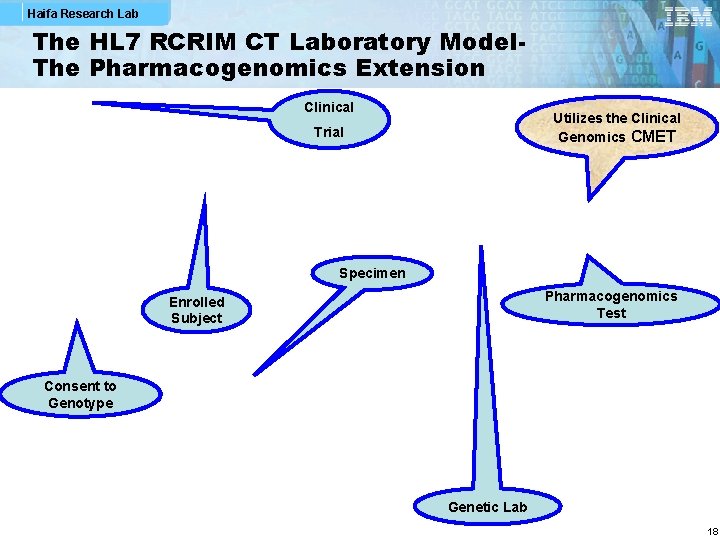
Haifa Research Lab The HL 7 RCRIM CT Laboratory Model. The Pharmacogenomics Extension Clinical Utilizes the Clinical Genomics CMET Trial Specimen Pharmacogenomics Test Enrolled Subject Consent to Genotype Genetic Lab 18

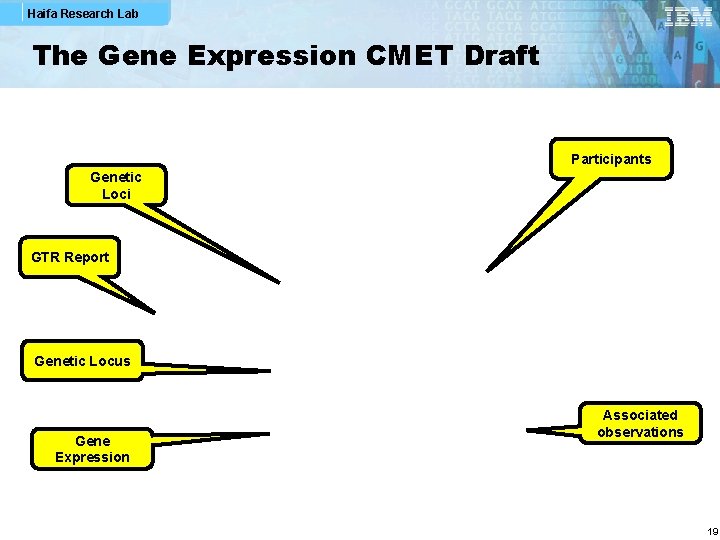
Haifa Research Lab The Gene Expression CMET Draft Participants Genetic Loci GTR Report Genetic Locus Gene Expression Associated observations 19

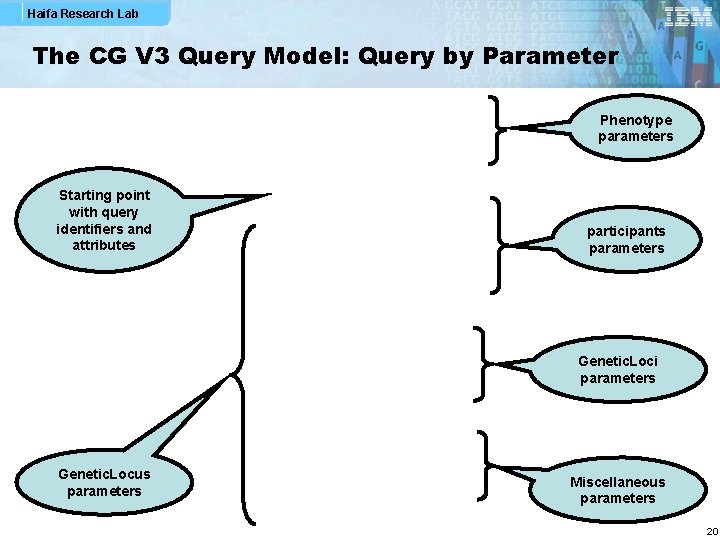
Haifa Research Lab The CG V 3 Query Model: Query by Parameter Phenotype parameters Starting point with query identifiers and attributes participants parameters Genetic. Loci parameters Genetic. Locus parameters Miscellaneous parameters 20

Haifa Research Lab V 2 Implementation Guides § The IG “Genetic Test Result Reporting to EHR” passed informative ballot § It is modeled after the HL 7 Version 2. 5. 1 Implementation Guide: Orders And Observations; Interoperable Laboratory Result Reporting To EHR (US Realm), Release 1 § Is used in a pilot of information exchange between Partners Healthcare and Intermountain Health Care 21

Haifa Research Lab V 2 update for January 2012 § Genetic variations § Cytogenetics 22

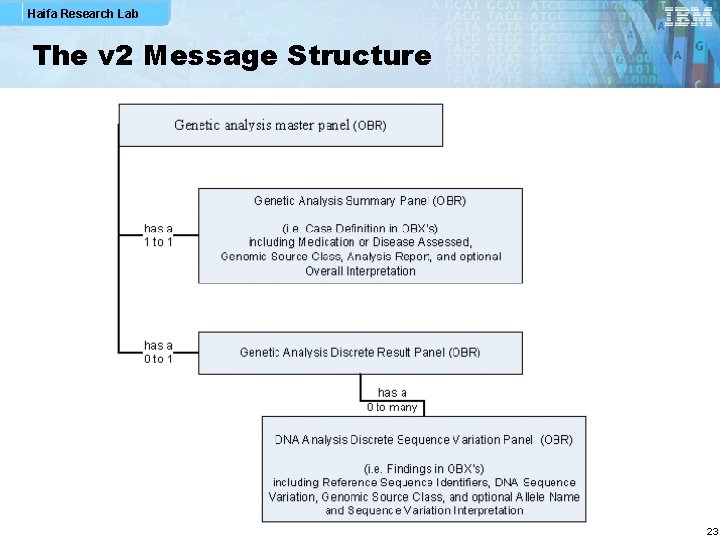
Haifa Research Lab The v 2 Message Structure 23

Haifa Research Lab V 2 Sample Message § OBR|1||PM-08 -J 00094^HPCGG- LMM^2. 16. 840. 1. 113883. 3. 167. 1^ISO|lm_DCM-pnl. B_L^Dilated Cardiomyopathy Panel B (5 genes)^99 LMM-ORDER-TESTID||20080702000000|20080702100909|||||234567891^Pump^Patrick^^^^^^ NPI^L||||||20080703000000|||F||||||00000009^Cardiovascular^99 HPCGG-GVIE -INDICATION^^^^^^Clinical Diagnosis and Family History of DCM|&Geneticist&Gene&&&&&NPI^^^^^^^HPCGGLMM&2. 16. 840. 1. 113883. 3. 167. 1&ISO||||||||55233 -1^Genetic analysis master panel ^LN § SPM|1|||119273009&Peripheral blood&SNM 3&&&&0707 Intl&&Blood, Peripheral|||||||20080702000000 § OBR|2||PM-08 -J 00094 -1^HPCGG- LMM^2. 16. 840. 1. 113883. 3. 167. 1^ISO|55232 -3^Genetic analysis summary panel^LN|||20080702000000||||||||20080703000000|||F||||^PM-08 J 00094&HPCGG-LMM&2. 16. 840. 1. 113883. 3. 167. 1&ISO § OBX|1|CWE|51967 -8^Genetic disease assessed^LN||399020009^DCM- Dilated Cardiomyopathy^SNM 3^^^0707 Intl||||||F|20080702100909||||||Laboratory for Molecular Medicine^L^22 D 1005307^^^CLIA&2. 16. 840. 1. 113883. 4. 7&ISO|1000 Laboratory Lane^Ste. 123^Cambridge^MA^99999^USA^B 24

Haifa Research Lab CDA IG: Genetic Testing Report (GTR) § Define an implementation guide for a genetic testing report that is both human readable and machine-processable § § Target at all types of GTR producers, e. g. , genetic labs, clin. geneticists Readable content is larger in scope E. g. , detailed description of the tests performed along with references Machine-processable should be limited, e. g. , exclude raw data § Ballot a Universal IG; then derive specific types of GTR: § Healthcare & Research § Realm-specific guides § Omic-specific guides § Developed using the MDHT open source tool (OHT) 25

Haifa Research Lab GTR - Design Principles § Follow existing report formats commonly used in healthcare & research § Emphasize interpretations & recommendations § Provide general background information on tests performed § Reference HL 7 Clinical Genomics instances (e. g. , v 3 or v 2 Genetic. Variation and Pedigree) as the place holders of full-blown raw genomic data and fully-structured family history data § Utilize patterns of ‘genotype-phenotype’ associations in the HL 7 v 3 Clinical Genomics Domain § Implement them as ‘clinical genomic statement’ entry-level templates (see next slide), enabling meaningful use of the data 26

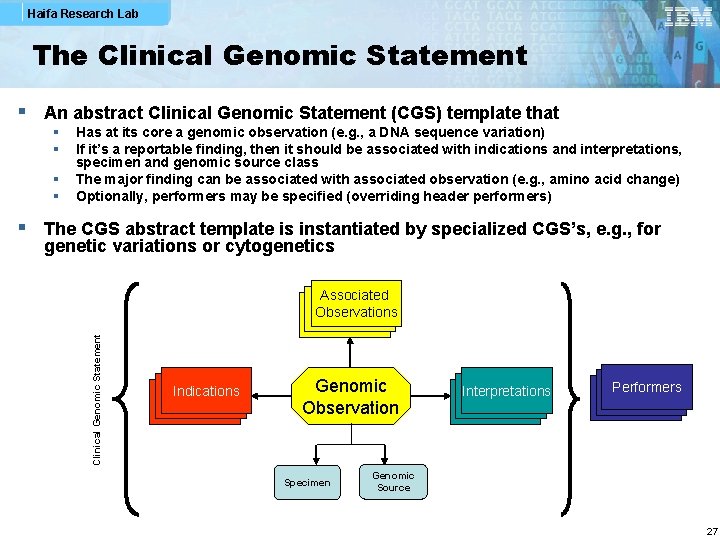
Haifa Research Lab The Clinical Genomic Statement § An abstract Clinical Genomic Statement (CGS) template that § § Has at its core a genomic observation (e. g. , a DNA sequence variation) If it’s a reportable finding, then it should be associated with indications and interpretations, specimen and genomic source class The major finding can be associated with associated observation (e. g. , amino acid change) Optionally, performers may be specified (overriding header performers) § The CGS abstract template is instantiated by specialized CGS’s, e. g. , for genetic variations or cytogenetics Clinical Genomic Statement Associated Observations Indications Genomic Observation Specimen Interpretations Performers Genomic Source 27

Haifa Research Lab Narrative and Structured Data § All CGS structured data items shall be part of clinical genomic statement (CGS) instances so that parsing applications can find the full semantics explicitly represented in one coherent structure § In the case of the overall interpretation, it is part of CGS that has references to the various testing interpretations § Sub-sections such as Indications, Interpretations and Specimen are mainly for presenting narrative, but they may also contain structured data § In this way, it is possible to have less redundant documents, e. g. , in the case where all tests reported in a GTR document have the same indication, an Indications section in the Summary section consists of a full-blown indication observation which all CGS indication observations reference § CGS structured data may point to the respective narrative in subsections (by means of XML ID) 28

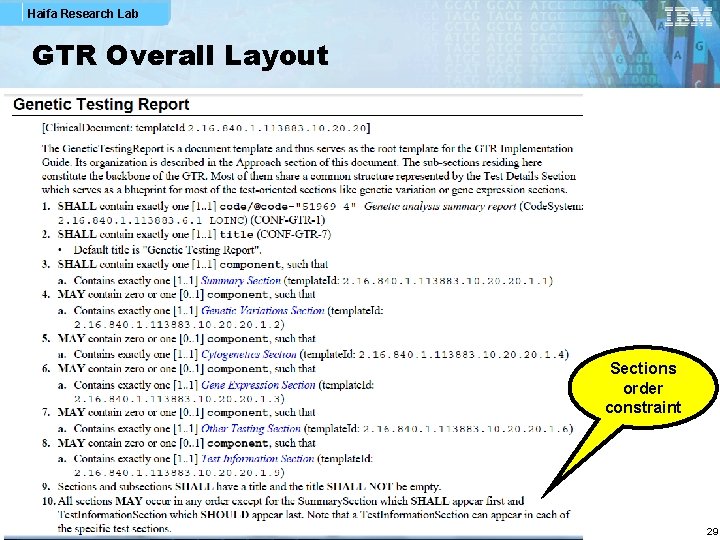
Haifa Research Lab GTR Overall Layout Sections order constraint 29

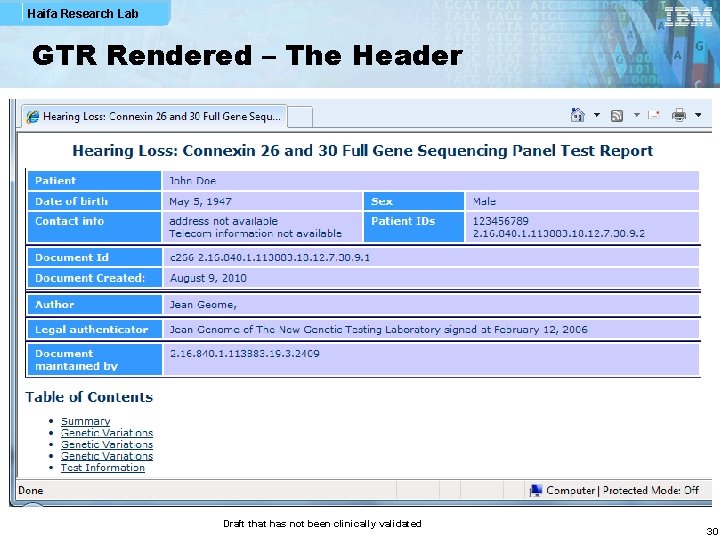
Haifa Research Lab GTR Rendered – The Header Draft that has not been clinically validated 30

Haifa Research Lab GTR Rendered – Summary Section Draft that has not been clinically validated 31

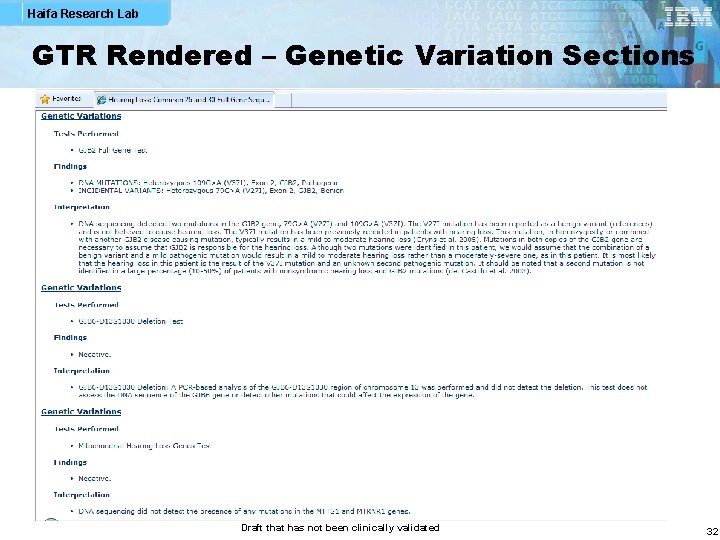
Haifa Research Lab GTR Rendered – Genetic Variation Sections Draft that has not been clinically validated 32

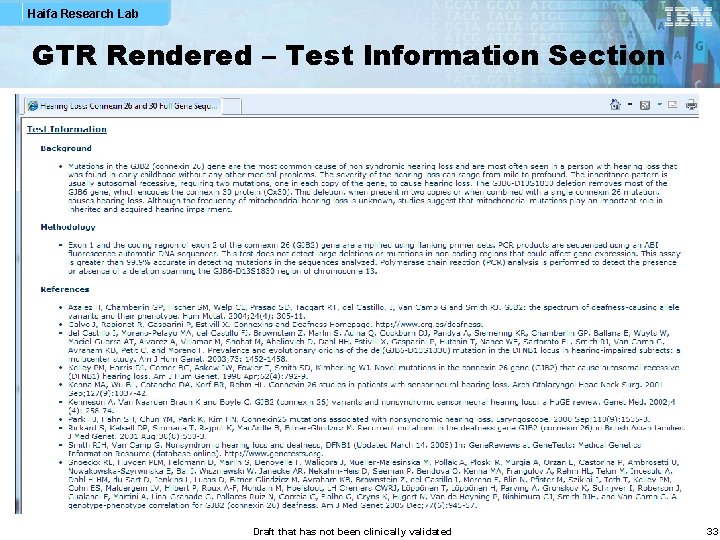
Haifa Research Lab GTR Rendered – Test Information Section Draft that has not been clinically validated 33

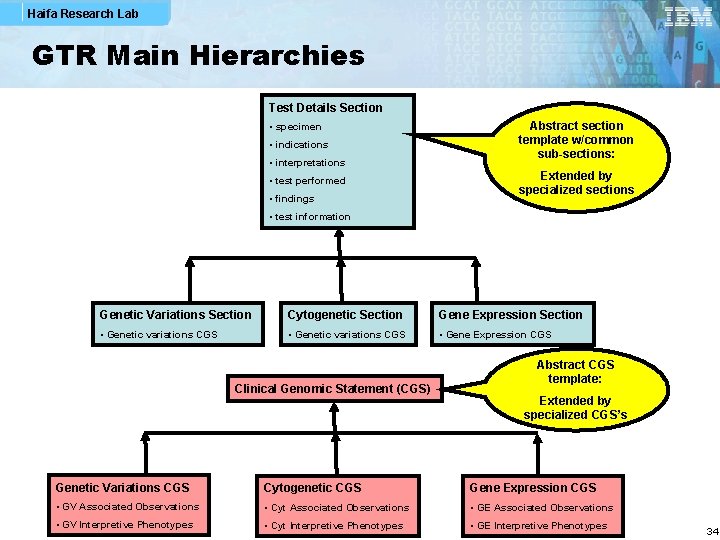
Haifa Research Lab GTR Main Hierarchies Test Details Section • specimen • indications • interpretations • test performed • findings Abstract section template w/common sub-sections: Extended by specialized sections • test information Genetic Variations Section Cytogenetic Section Gene Expression Section • Genetic variations CGS • Gene Expression CGS Clinical Genomic Statement (CGS) Abstract CGS template: Extended by specialized CGS’s Genetic Variations CGS Cytogenetic CGS Gene Expression CGS • GV Associated Observations • Cyt Associated Observations • GE Associated Observations • GV Interpretive Phenotypes • Cyt Interpretive Phenotypes • GE Interpretive Phenotypes 34

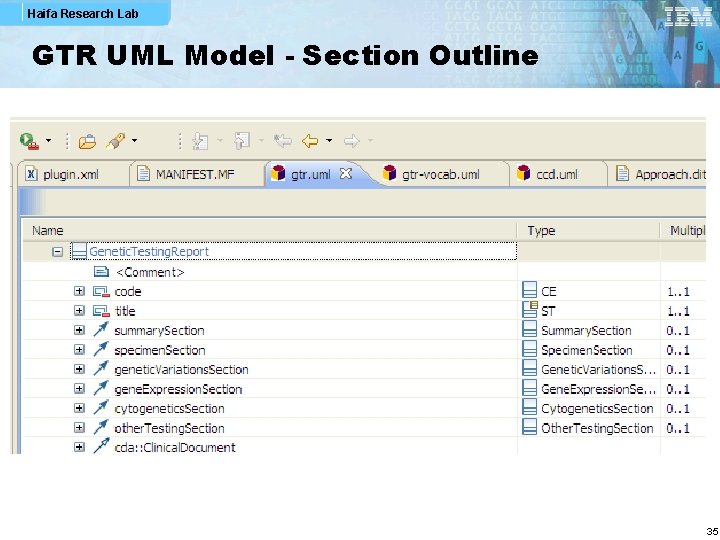
Haifa Research Lab GTR UML Model - Section Outline 35

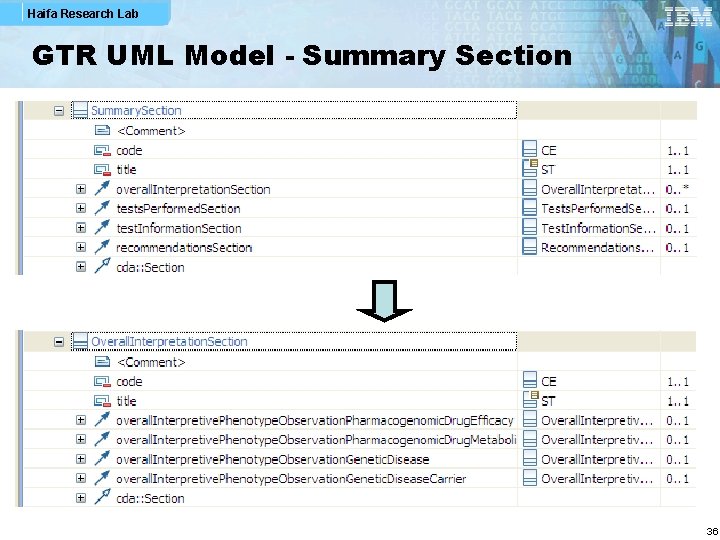
Haifa Research Lab GTR UML Model - Summary Section 36

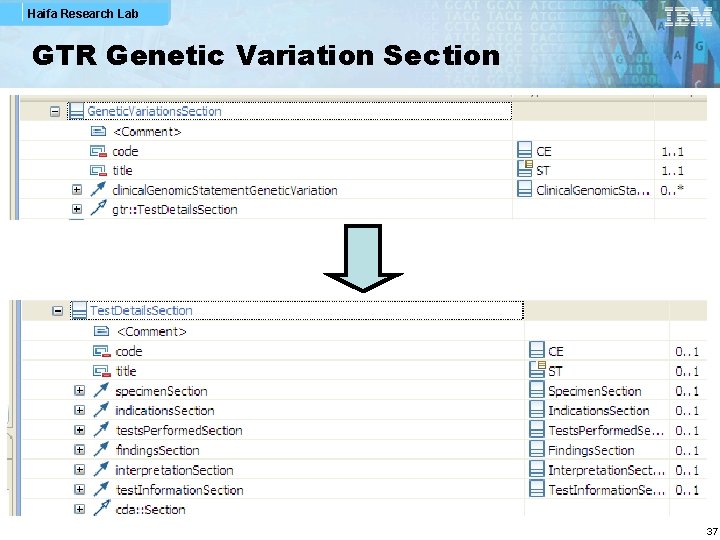
Haifa Research Lab GTR Genetic Variation Section 37

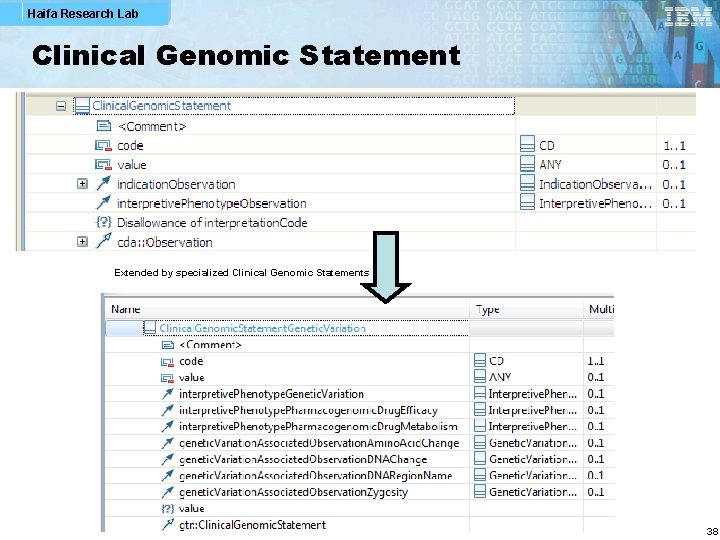
Haifa Research Lab Clinical Genomic Statement Extended by specialized Clinical Genomic Statements 38

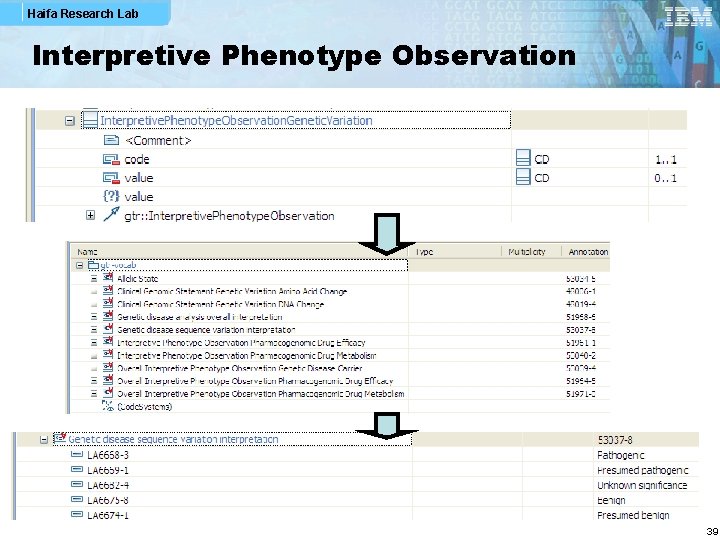
Haifa Research Lab Interpretive Phenotype Observation 39

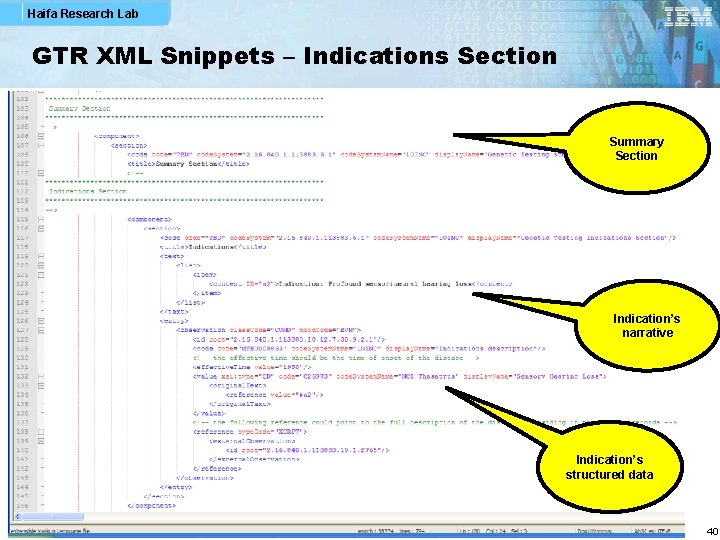
Haifa Research Lab GTR XML Snippets – Indications Section Summary Section Indication’s narrative Indication’s structured data 40

Haifa Research Lab GTR XML Snippets – Specimen Section Specimen’s narrative Specimen’s structured data 41

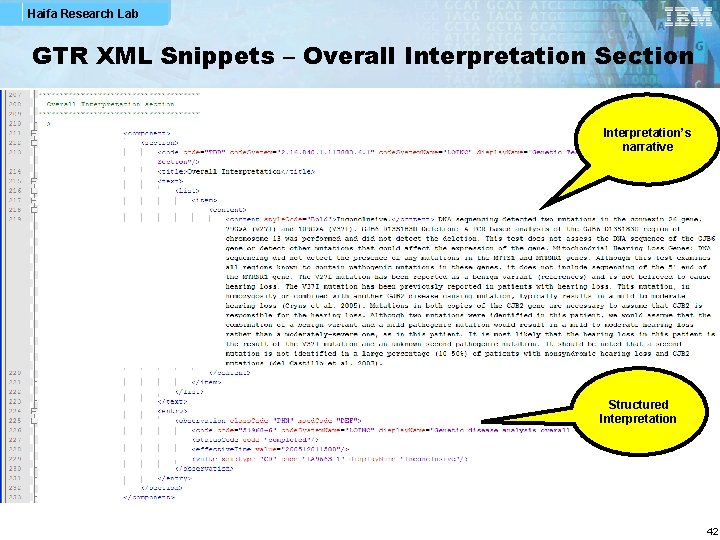
Haifa Research Lab GTR XML Snippets – Overall Interpretation Section Interpretation’s narrative Structured Interpretation 42

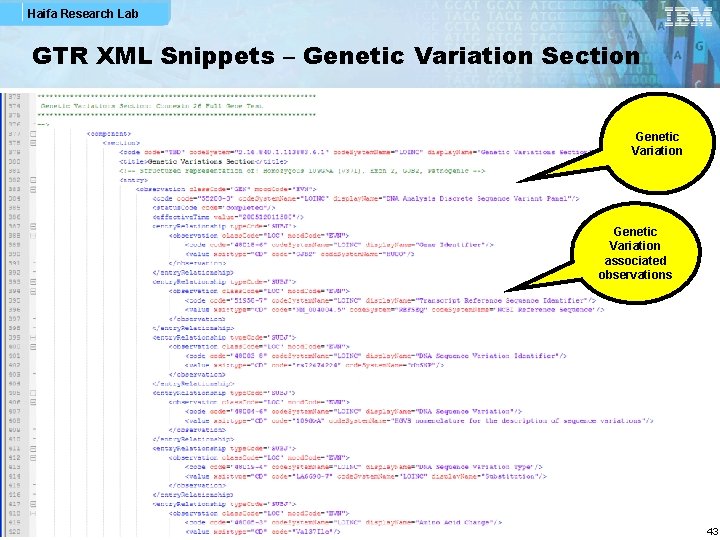
Haifa Research Lab GTR XML Snippets – Genetic Variation Section Genetic Variation associated observations 43

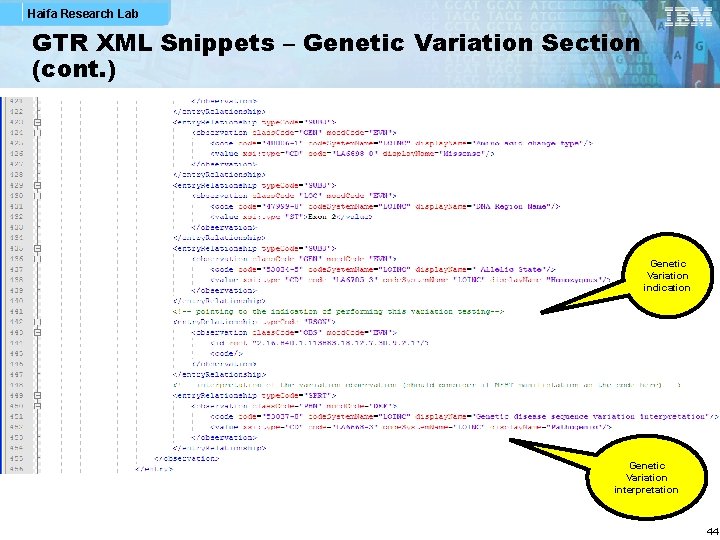
Haifa Research Lab GTR XML Snippets – Genetic Variation Section (cont. ) Genetic Variation indication Genetic Variation interpretation 44

Haifa Research Lab CDA GTR Ballot Status § Balloted as DSTU and passed in October 2010 § Still under ballot to refine & reconcile ballot comments § Main issues: § Vocabulary: § Universal spec vs. Realm (e. g. mandate the use of LOINC code? ) § Binding syntax (align with new vocabulary spec and the respective SDWG guidance for CDA IGs) § Layout: § Semantics – compare to recommended layouts in the literature § Syntactic – works closely with MDHT developers to adhere to SDWG guidelines § Sections specific to every type of genetic test (derived from abstract) § Section and Entry level template ids registration (when layout agreed) § Suggestion to add drug safety template (considered for future use) 45

Haifa Research Lab Alignment Among the Various Specs § v 3 specs and CDA are all based the RIM § CDA GTR-IG will be based on CDA R 3 § Depending on the “right side” of R 3, if it allows RIM-based domain models, then alignment is trivial § v 3 -v 2 alignment: § Proposal: represent semantics with v 3 and implement it in various ways, one of which is v 2; develop an “v 2 ITS” for the v 3 models § See proposal made by Amnon in a separate presentation (click here to see that presentation) 46

Haifa Research Lab Utilizations in HL 7 § Clinical Trials: HL 7 RCRIM Work Group (clinical trials specs) utilized the CG DSTU model (Genetic Locus) in their Pharmacogenomics message, which was an extension of the CTLab message (an approved but expired DSTU) § Laboratory: The Lab Work Group might utilize a constrained version of the Genetic Variation model in their next release if the Lab Result message 47

Haifa Research Lab Selected Implementation § v 2 § Exchange of genetic testing results between Intermountain and Harvard § v 3 § The Family History spec is used in Mass General Hospital § Expanding to other family history applications including the US Surgeon General Family History tool § The Genetic Variation model is used in Hypergenes (a European project on essential hypertension, http: //www. hypergenes. eu/) § The Pedigree and Genetic Variation models are used in Italy, the Rizzoli institute in Bologna § CDA § GTR has been used in u. Health – a PHR/EHR system in Korea 48

Haifa Research Lab HL 7 WG Health Check – Need to Improve! § § § § Active projects SWOT current 3 year plan current Mission and charter current Co-chair post-WGM survey participation Ballot presence Minutes posted since last WGM Last listserv activity Wiki presence WG conference calls schedules Steering division conference call participation Steering division co-chair (TSC representation) election participation WG rep at steering division WGM WG meetings at WGM scheduled WG has an approved DMP based on review of the updated template 49

Haifa Research Lab Planning ahead § May 2012 WGM (Vancouver) § Schedule (from Tuesday Q 3 to Thursday Q 2) § Joint meetings § AP – Wed Q 4 § CDS - informal § OO+AP – Wed Q 1 § Weekly conf. calls § Continue Tuesday’s 11 EST § Submit ‘renewed’ PSSs for GTR and Omics § Prepare to ballot GTR, Omics & Sequencing storyboards in May 2012 50

Haifa Research Lab Summary § Small group coping with § Various HL 7 formats: v 3, v 2 and CDA § Clinical & Research environments § Developing a DAM and component models (CMETs) to be used in other HL 7 domains § Genetic Variation § Gene Expression § CDA Genetic Testing Report (GTR) § Bridge from raw data to human readable reports and bubbled-up data § Model-driven development of standards (use of MDHT CDA Editor) 51

Haifa Research Lab The End • Thank you for your attention… • Questions? Contact Amnon at shabo@il. ibm. com • Comments of general interest should be posted to the CG mailing list at clingenomics@lists. hl 7. org 52