HL 7 Clinical Genomics RIM Constraining Issues May





- Slides: 17

HL 7 Clinical Genomics – RIM Constraining Issues May 2008 The HL 7 Clinical Genomics SIG Amnon Shabo (Shvo), Ph. D HL 7 Clinical Genomics SIG Co-chair and Modeling Facilitator HL 7 Structured Documents TC CDA R 2 Co-editor CCD Implementation Guide Co-editor

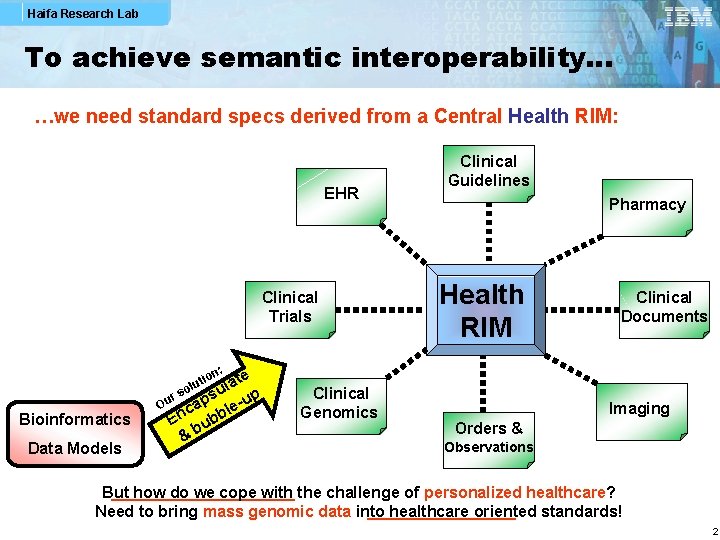
Haifa Research Lab To achieve semantic interoperability… …we need standard specs derived from a Central Health RIM: EHR Clinical Trials Bioinformatics Data Models Pharmacy Health RIM Clinical Documents n: te a l p su r Ou cap le-u En ubb b & so io lut Clinical Guidelines Clinical Genomics Imaging Orders & Observations But how do we cope with the challenge of personalized healthcare? Need to bring mass genomic data into healthcare oriented standards! 2

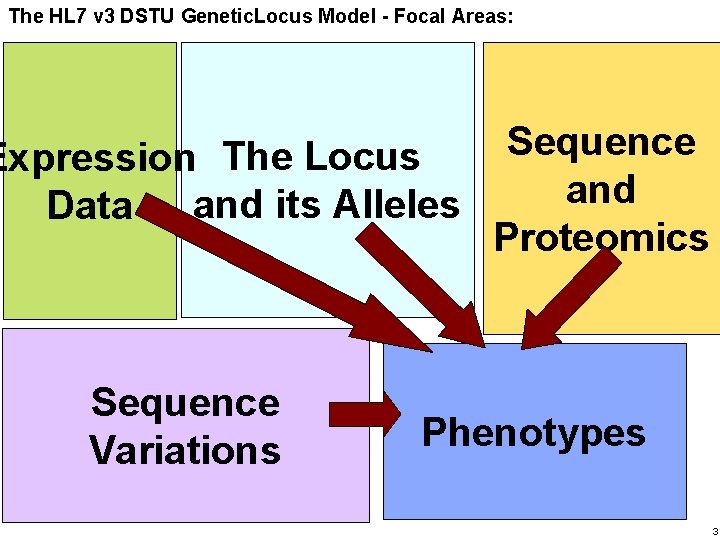
Haifa Research The HL 7 v 3 Lab DSTU Genetic. Locus Model - Focal Areas: Sequence Expression The Locus and its Alleles Data Proteomics Sequence Variations Phenotypes 3

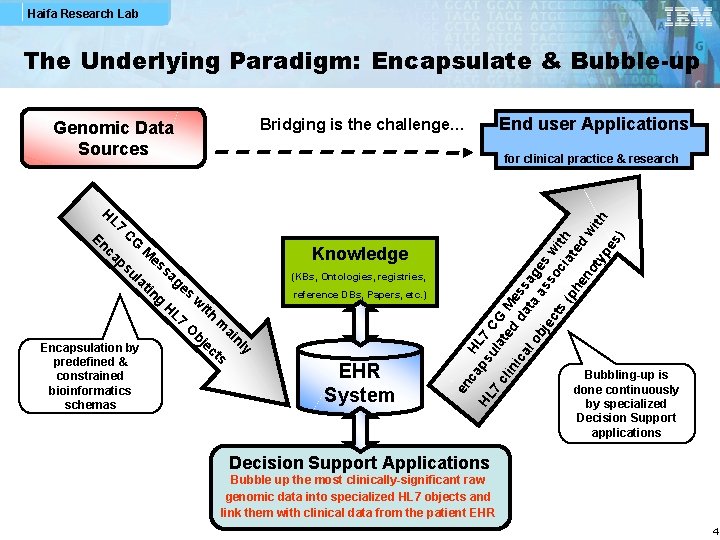
Haifa Research Lab The Underlying Paradigm: Encapsulate & Bubble-up (KBs, Ontologies, registries, reference DBs, Papers, etc. ) EHR System HL Knowledge en c En CG ca M es ps sa ul ge at in s g w H L 7 ith m O ai bj nl e y Encapsulation by ct s ap HL su 7 C 7 la G cl te M in d ic da ess al ta ag ob as es je so w ct s ci ith (p at e he no d w ith ty pe s) for clinical practice & research H L 7 predefined & constrained bioinformatics schemas End user Applications Bridging is the challenge… Genomic Data Sources Bubbling-up is done continuously by specialized Decision Support applications Decision Support Applications Bubble up the most clinically-significant raw genomic data into specialized HL 7 objects and link them with clinical data from the patient EHR 4

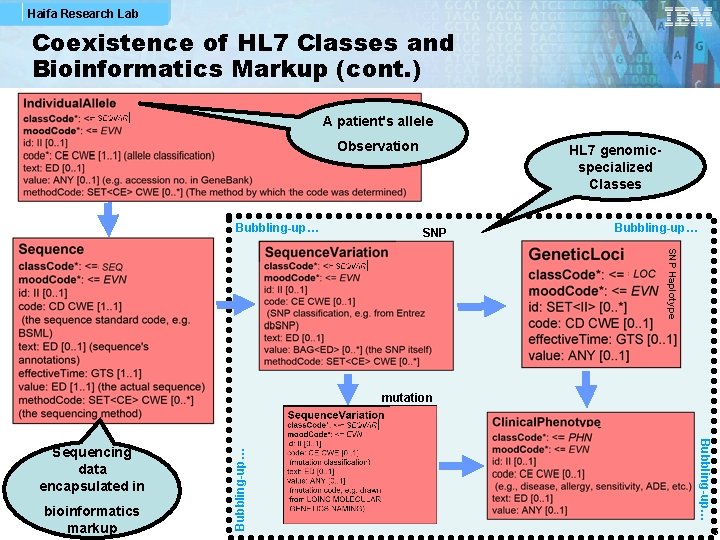
Haifa Research Lab Coexistence of HL 7 Classes and Bioinformatics Markup (cont. ) A patient's allele Observation Bubbling-up… HL 7 genomicspecialized Classes SNP Bubbling-up… SNP Haplotype bioinformatics markup Bubbling-up… Sequencing data encapsulated in Bubbling-up… mutation 5

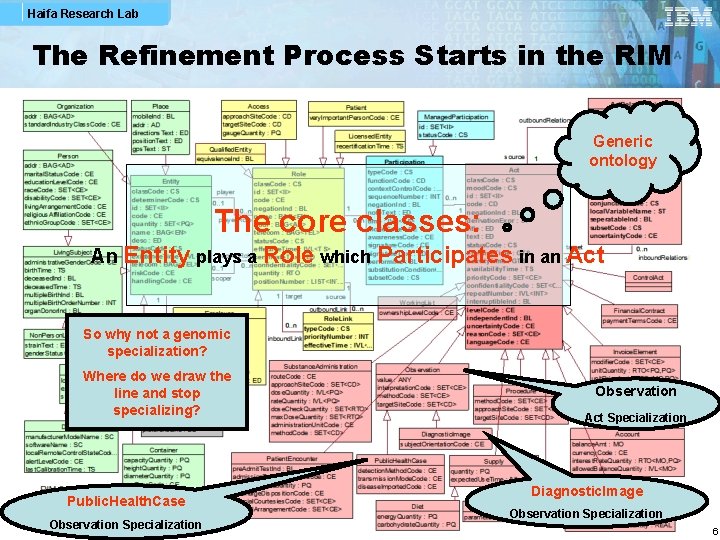
Haifa Research Lab The Refinement Process Starts in the RIM Generic ontology The core classes: An Entity plays a Role which Participates in an Act So why not a genomic specialization? Where do we draw the line and stop specializing? Public. Health. Case Observation Specialization Observation Act Specialization Diagnostic. Image Observation Specialization 6

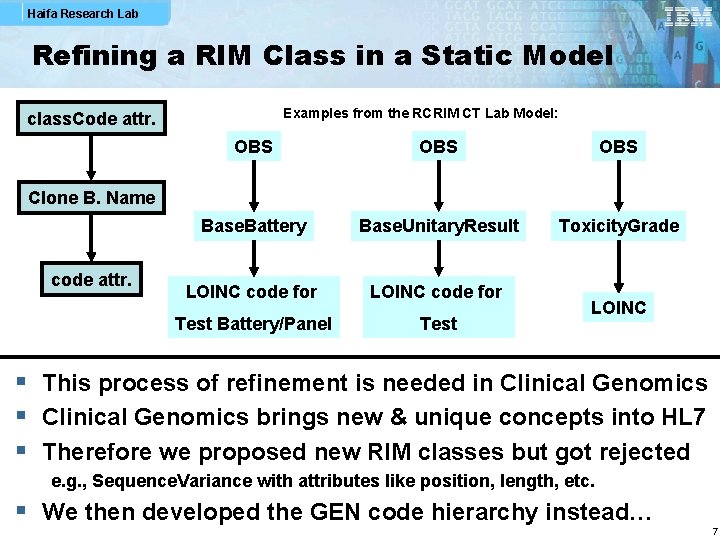
Haifa Research Lab Refining a RIM Class in a Static Model Examples from the RCRIM CT Lab Model: class. Code attr. OBS OBS Base. Battery Base. Unitary. Result Toxicity. Grade LOINC code for Test Battery/Panel Test Clone B. Name code attr. LOINC § This process of refinement is needed in Clinical Genomics § Clinical Genomics brings new & unique concepts into HL 7 § Therefore we proposed new RIM classes but got rejected e. g. , Sequence. Variance with attributes like position, length, etc. § We then developed the GEN code hierarchy instead… 7

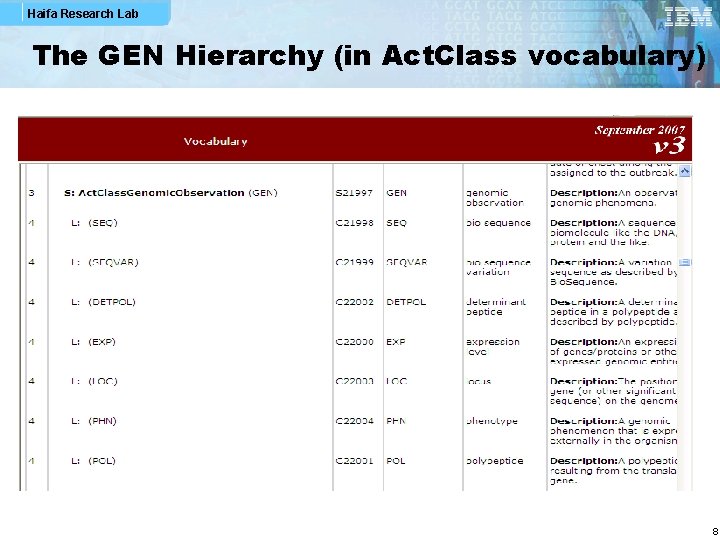
Haifa Research Lab The GEN Hierarchy (in Act. Class vocabulary) 8

Haifa Research Lab The GEN Hierarchy (in Act. Class vocabulary) § The GEN code hierarchy was accepted by RIM Harmonization already in July 2006! § It indentifies core classes of the genomic models § Enables further specialization of classes like sequence variation by using the code attribute § It allows Decision Support & Data Mining to bubble-up, § i. e. , creating and looking for genotype phenotype associations § We need minor refinements of this hierarchy, e. g. : § Add ALLELE code to the GEN hierarchy § This is also addressing Bob Dolin’s ballot comment about SEQVAR for Individual. Allele wild type which is not an appropriate code § PHN (Phenotype) is a genomic-related observation (edit GEN definition) 9

Haifa Research Lab Complex Genotype Phenotype Associations “Some patients, particularly young children, may be born with a genetic mutation which means they are at risk of hearing loss after taken antibiotics called aminoglycosides. There is now a drive to consider screening patients for the genetic mutation known as m. 1555 A-G which is held in around 1 in 1, 611 newborns in the USA, 1 in 206 newborns in New Zealand 1 in 40, 000 newborns in the UK. Aminoglycosides are valuable antibiotics used for serious infections such as complicated urinary tract infections, TB and septicemia. They are known to potentially cause damage to the ear - otoxicity. Individuals holding the mutation have an inherited predisposition which makes them extremely sensitive to the effects - they can end up with severe and permanent hearing loss". 10

Haifa Research Lab The PHN (Phenotype) Class Code § Signifies that an observation is a phenotype § Typically is associated with a source genomic observation § But not necessarily in cases where the exact genetic sources is yet unknown § A genomic observation could be associated with other genomic observations or with other observations not necessarily classified as phenotypes § Prepare the ground for a ‘disease model’ expressed in RIM so that patient data could be better checked against it § The IBM Clinical Genomics solution as used the Hypergenes project - http: //www. hypergenes. eu/) relies on the GEN hierarchy and performs semantic computations based on these codes 11

Haifa Research Lab General Vocabulary Issues § What are the clear-cut criteria that distinguish between Act. Class and Act. Code, preferably in a way that a designer of a clinical decision support application could use them programmatically? § Act. Code should specialize Act. Class but current guidance and the actual values in the two vocabularies sometime blur the differences between the two, e. g. : § _Medication. Observation. Type act code doesn’t specialize any act class § Specimen. Transport act code could be specializing TRANS or SPCTRT § _Act. Cognitive. Professional. Service. Code act code doesn’t specialize a. class § If indeed one specializes the other, why not merge them into one coherent vocabulary where sub-typing will become explicit? § The fields class. Code, [clone name] and code will bind to the appropriate layer in this proposed consolidated vocabulary 12

Haifa Research Lab What if GEN is Moved to Act. Code… § In lack of class code & clone name - need to rely on the code attribute, however… § This poses the following problems: § Code attribute is often drawn from external terminology § Giving up class code & clone name leads to loss of granularity in the data represented by the class § Inferring the generic GEN code from a value in the code attribute might not be easy, especially if drawn from external terminology 13

Haifa Research Lab Clone Business Names DO Carry Semantics § Current guidance: Clone business names should not carry semantics § Rebuttal: These names are meaningful to committee members and are balloted and approved by the HL 7 membership ? ? ? § An example – CDA Authoring. Device vs. Device § The CDA R 2 Authoring. Device and Device clones have the same class code (DEV) and the same binding of the code attribute (Entity. Code). § In fact, they have the very same set of attributes refined exactly the same (cardinality, coding strength, vocab, etc). § The only way to distinguish between the two is through the clone name (unless you rely on the traversal path). § The two clones are semantically distinct: Authoring. Device has to do with the authoring of the document and Device has to do with some specimen mentioned in the body of the document. § In addition, the clone Authoring. Device does carry semantics which cannot be found in either Entity. Class or Entity. Code vocabularies. 14

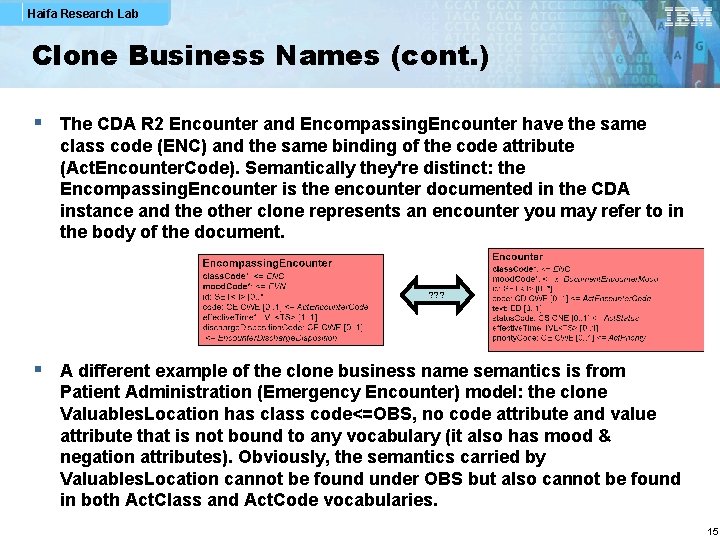
Haifa Research Lab Clone Business Names (cont. ) § The CDA R 2 Encounter and Encompassing. Encounter have the same class code (ENC) and the same binding of the code attribute (Act. Encounter. Code). Semantically they're distinct: the Encompassing. Encounter is the encounter documented in the CDA instance and the other clone represents an encounter you may refer to in the body of the document. ? ? ? § A different example of the clone business name semantics is from Patient Administration (Emergency Encounter) model: the clone Valuables. Location has class code<=OBS, no code attribute and value attribute that is not bound to any vocabulary (it also has mood & negation attributes). Obviously, the semantics carried by Valuables. Location cannot be found under OBS but also cannot be found in both Act. Class and Act. Code vocabularies. 15

Haifa Research Lab Formalize Clone Business Names Or… § Either… § Formalize the clone business name into a vocabulary domain, intertwined in the consolidated Act. Class/Ac. Code vocabulary § Adding a definition for each clone name is already done in the walkthrough of each model, and possibly in the glossary § In this way, we can relate to clone names as codes in the cascading 'identification' of a class § This could ease the burden of class identification by the class code attribute, if indeed the wish is to keep the Act. Class as minimal as possible § Or… § Disallow the use of clone business names as if they are class names § Enforce the automatic naming mechanism of clone names § In either of the options: § Continue to propagate clone names to the generated XML schemas! 16

Haifa Research Lab Thank You for Your Attention… § Questions? § Comments: shabo@il. ibm. com 17