HISTORY 1928 Frederick Griffith Performed bacterial transformation experiments

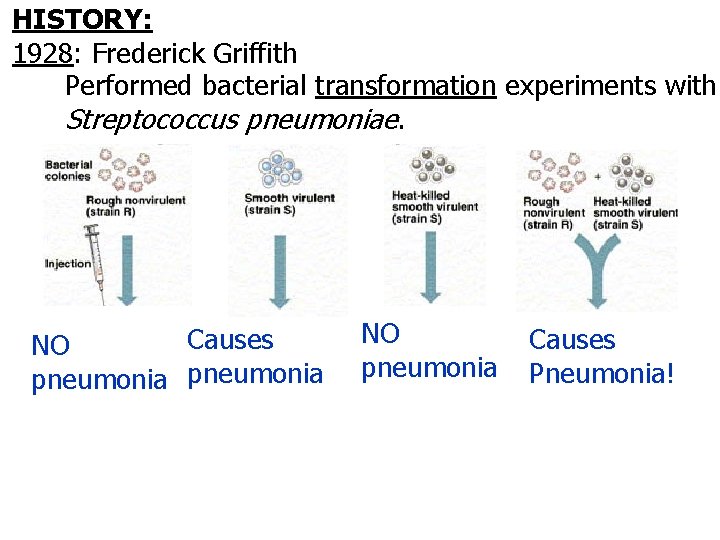
HISTORY: 1928: Frederick Griffith Performed bacterial transformation experiments with Streptococcus pneumoniae. Causes NO pneumonia Causes Pneumonia!
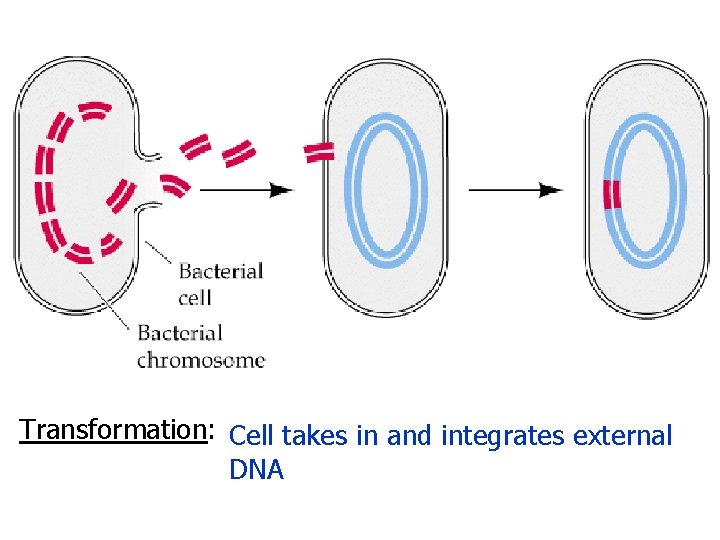
Transformation: Cell takes in and integrates external DNA
By 1940, Known information included: Chromosomes carry: heredity material Chromosomes are made of: DNA & protein Scientists assumed: protein was the genetic material
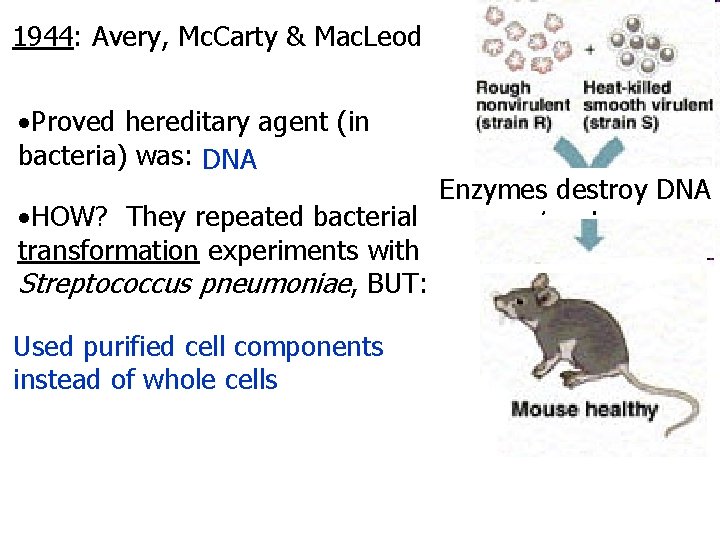
1944: Avery, Mc. Carty & Mac. Leod Proved hereditary agent (in bacteria) was: DNA HOW? They repeated bacterial transformation experiments with Streptococcus pneumoniae, BUT: Used purified cell components instead of whole cells Enzymes destroy lipids, Enzymes destroy DNA Enzymes destroy RNA carbs, and proteins
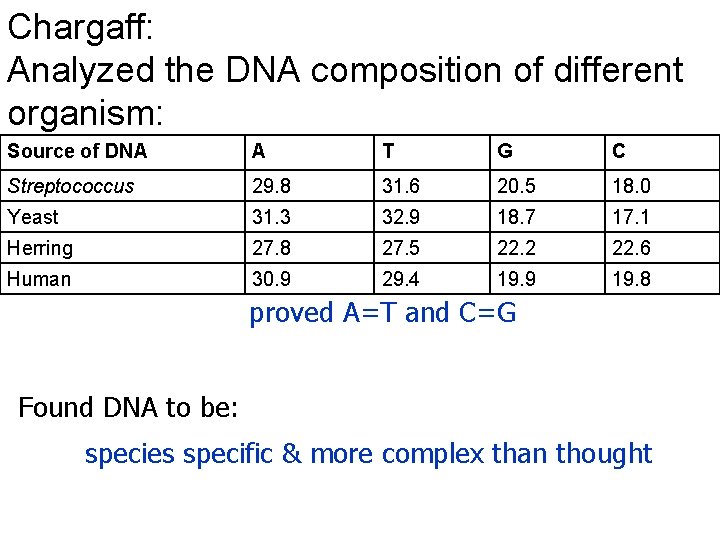
Chargaff: Analyzed the DNA composition of different organism: Source of DNA A T G C Streptococcus 29. 8 31. 6 20. 5 18. 0 Yeast 31. 3 32. 9 18. 7 17. 1 Herring 27. 8 27. 5 22. 2 22. 6 Human 30. 9 29. 4 19. 9 19. 8 proved A=T and C=G Found DNA to be: species specific & more complex than thought
1952: Hershey & Chase Conducted experiments with the T 2 bacteriophage (virus).
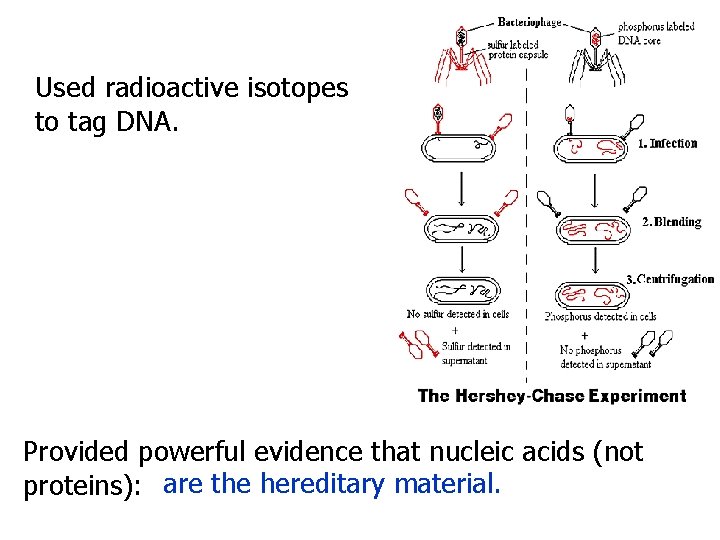
Used radioactive isotopes to tag DNA. Provided powerful evidence that nucleic acids (not proteins): are the hereditary material.

Sparked research by many famous scientists who contributed to the discovery of DNA’s structure and chemical makeup: Pauling, Wilkins, and x-ray diffraction Rosalind Franklin:

1953: Watson & Crick Published their model of DNA structure in Nature magazine. 3 minute clip with watson. Link on right side of page http: //www. pbs. org/wn et/dna/episode 1/index. h tml# Won Nobel prize in 1962: with Wilkins (Franklin had died at age 38)
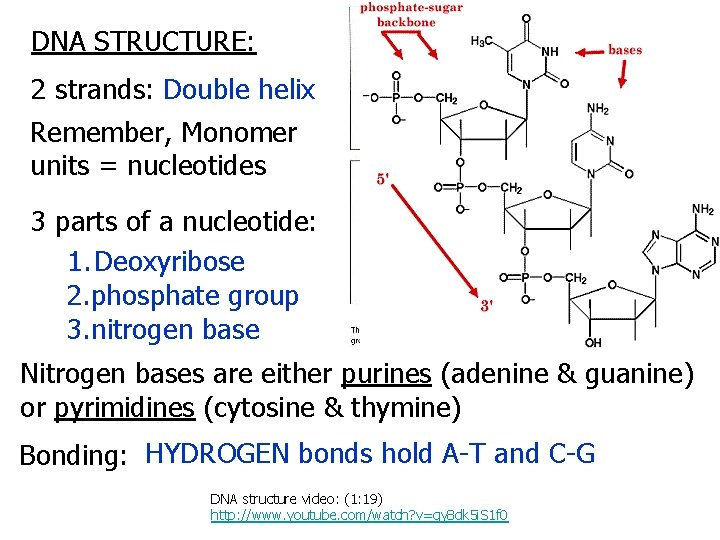
DNA STRUCTURE: 2 strands: Double helix Remember, Monomer units = nucleotides 3 parts of a nucleotide: 1. Deoxyribose 2. phosphate group 3. nitrogen base Nitrogen bases are either purines (adenine & guanine) or pyrimidines (cytosine & thymine) Bonding: HYDROGEN bonds hold A-T and C-G DNA structure video: (1: 19) http: //www. youtube. com/watch? v=qy 8 dk 5 i. S 1 f 0

DNA Replication: during S phase of interphase Fast Complex (See Figure 16. 9, p. 299 for the basic concept) Remarkably accurate: errors = 1/billion nucleotides
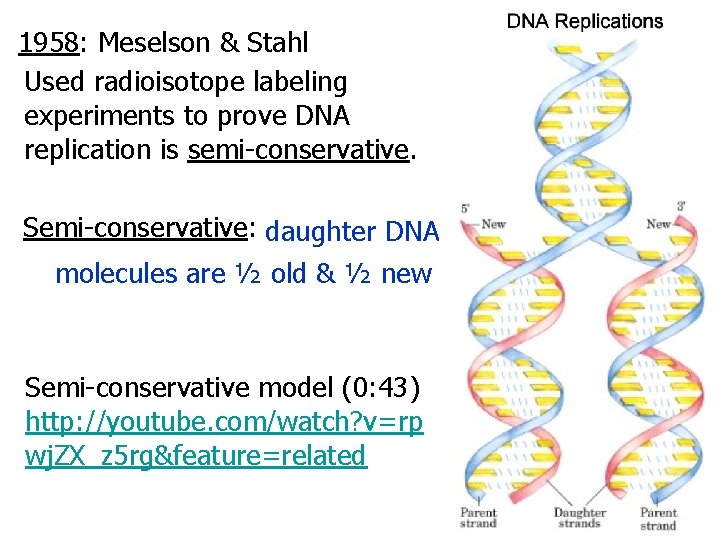
1958: Meselson & Stahl Used radioisotope labeling experiments to prove DNA replication is semi-conservative. Semi-conservative: daughter DNA molecules are ½ old & ½ new Semi-conservative model (0: 43) http: //youtube. com/watch? v=rp wj. ZX_z 5 rg&feature=related
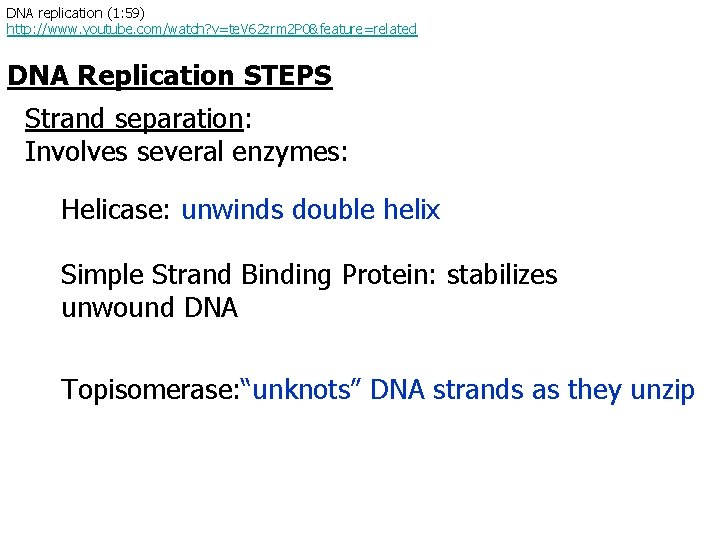
DNA replication (1: 59) http: //www. youtube. com/watch? v=te. V 62 zrm 2 P 0&feature=related DNA Replication STEPS Strand separation: Involves several enzymes: Helicase: unwinds double helix Simple Strand Binding Protein: stabilizes unwound DNA Topisomerase: “unknots” DNA strands as they unzip
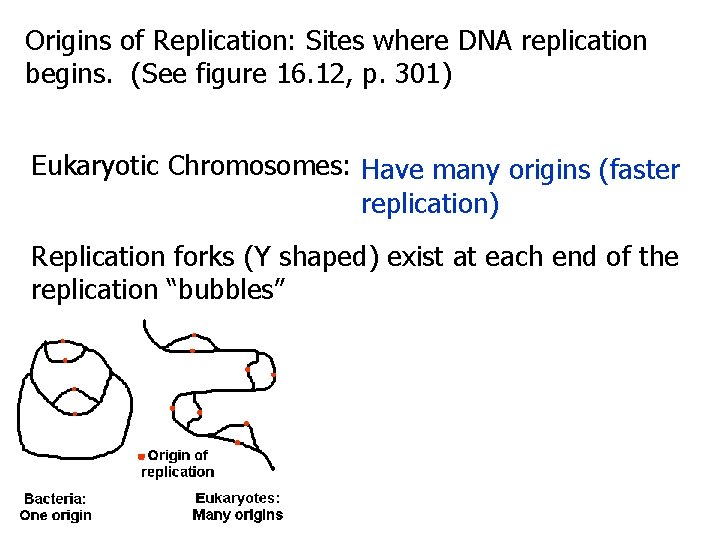
Origins of Replication: Sites where DNA replication begins. (See figure 16. 12, p. 301) Eukaryotic Chromosomes: Have many origins (faster replication) Replication forks (Y shaped) exist at each end of the replication “bubbles”
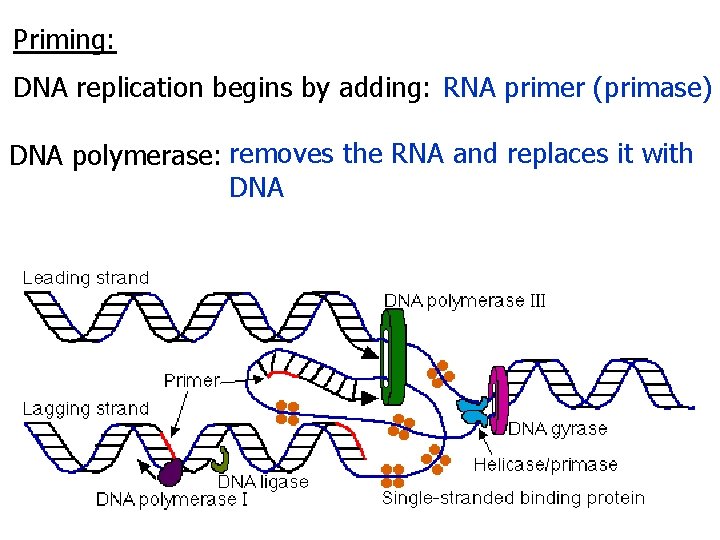
Priming: DNA replication begins by adding: RNA primer (primase) DNA polymerase: removes the RNA and replaces it with DNA
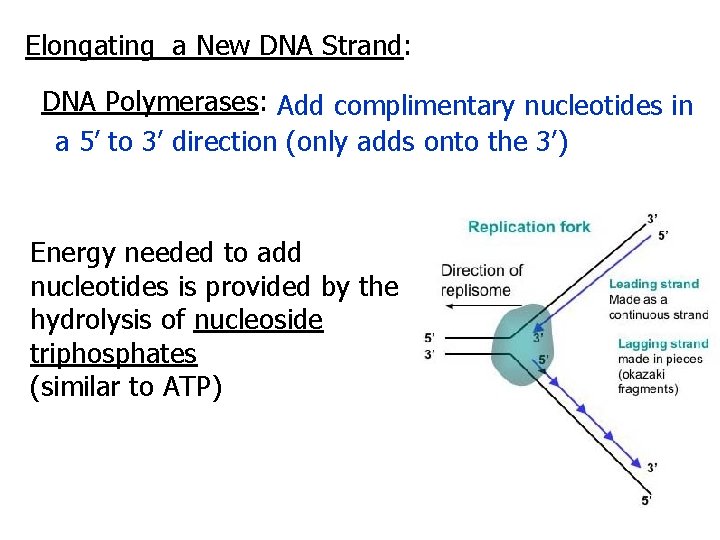
Elongating a New DNA Strand: DNA Polymerases: Add complimentary nucleotides in a 5’ to 3’ direction (only adds onto the 3’) Energy needed to add nucleotides is provided by the hydrolysis of nucleoside triphosphates (similar to ATP)
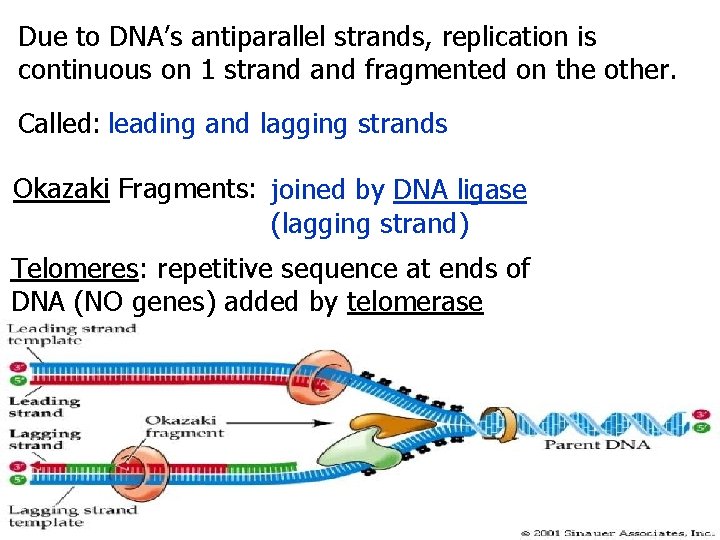
Due to DNA’s antiparallel strands, replication is continuous on 1 strand fragmented on the other. Called: leading and lagging strands Okazaki Fragments: joined by DNA ligase (lagging strand) Telomeres: repetitive sequence at ends of DNA (NO genes) added by telomerase

Proofreading and DNA repair: Initial pairing errors occur about 1/10, 000 Accidental DNA changes can be caused by: chemicals, radiation, x-rays, UV light. . . Most errors are repaired by: more than 130 kinds of proofreading enzymes (in humans)
HISTORY: 1909: Garrod Suggested genes dictate phenotypes through: enzymes (catalyzing reactions) 1930’s: Beadle & Ephrussi Speculated that Drosophila eye color mutations resulted from: missing enzyme 1942: Beadle & Tatum Used bread mold to demonstrate the relationship between: genes and enzymes Formulated the “one-gene-one-protein” hypothesis: each gene codes for a specific polypeptide (more accurate to say one gene = either polypeptide or RNA product)
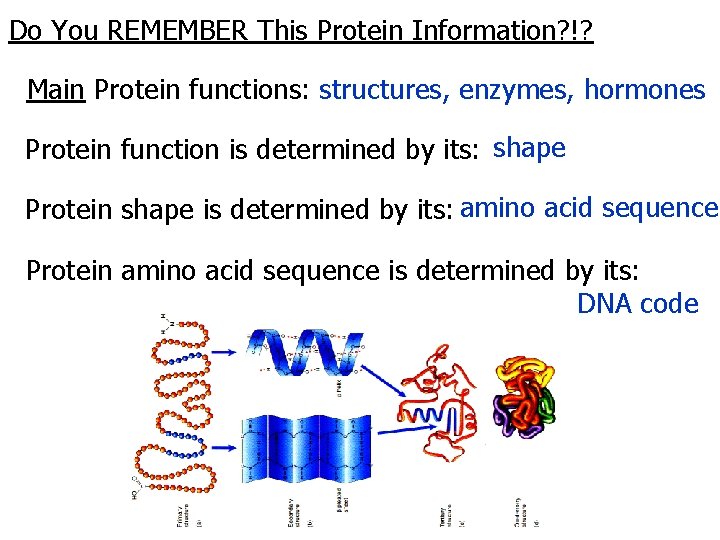
Do You REMEMBER This Protein Information? !? Main Protein functions: structures, enzymes, hormones Protein function is determined by its: shape Protein shape is determined by its: amino acid sequence Protein amino acid sequence is determined by its: DNA code
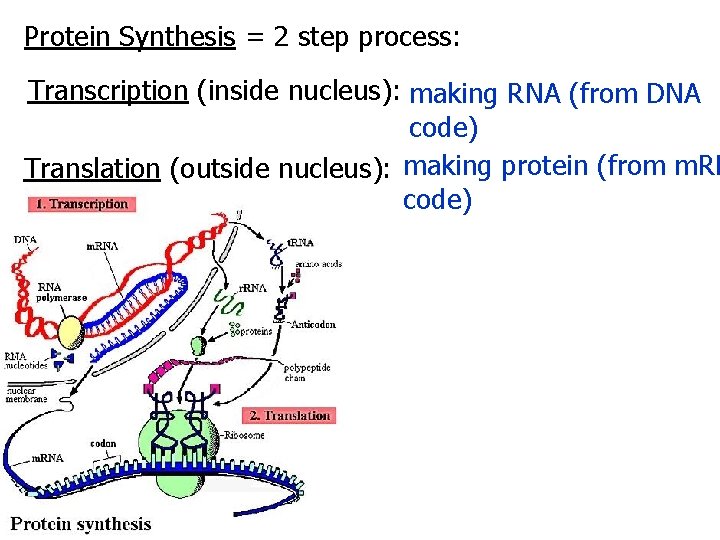
Protein Synthesis = 2 step process: Transcription (inside nucleus): making RNA (from DNA code) Translation (outside nucleus): making protein (from m. RN code)
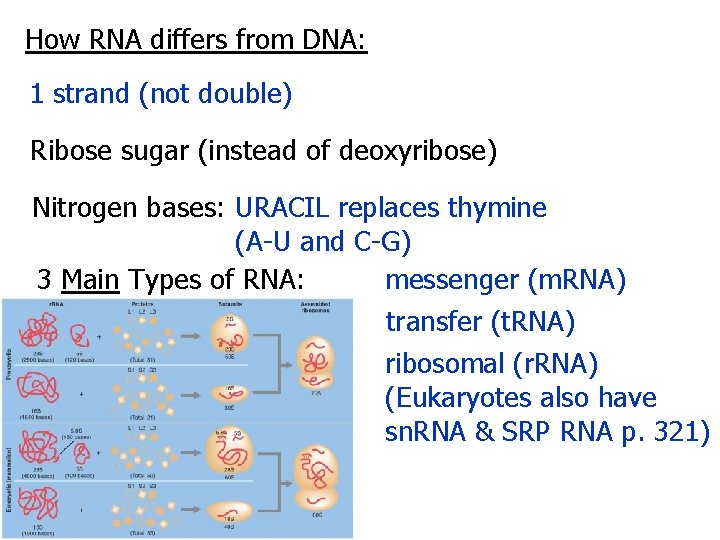
How RNA differs from DNA: 1 strand (not double) Ribose sugar (instead of deoxyribose) Nitrogen bases: URACIL replaces thymine (A-U and C-G) messenger (m. RNA) 3 Main Types of RNA: transfer (t. RNA) ribosomal (r. RNA) (Eukaryotes also have sn. RNA & SRP RNA p. 321)
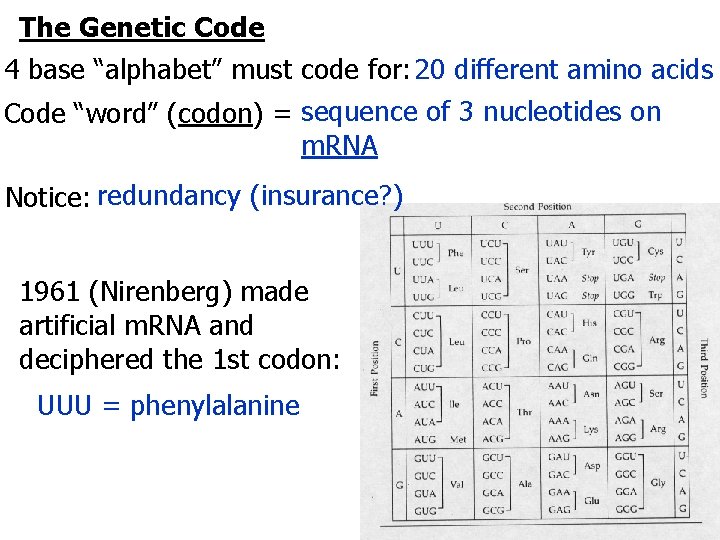
The Genetic Code 4 base “alphabet” must code for: 20 different amino acids Code “word” (codon) = sequence of 3 nucleotides on m. RNA Notice: redundancy (insurance? ) 1961 (Nirenberg) made artificial m. RNA and deciphered the 1 st codon: UUU = phenylalanine
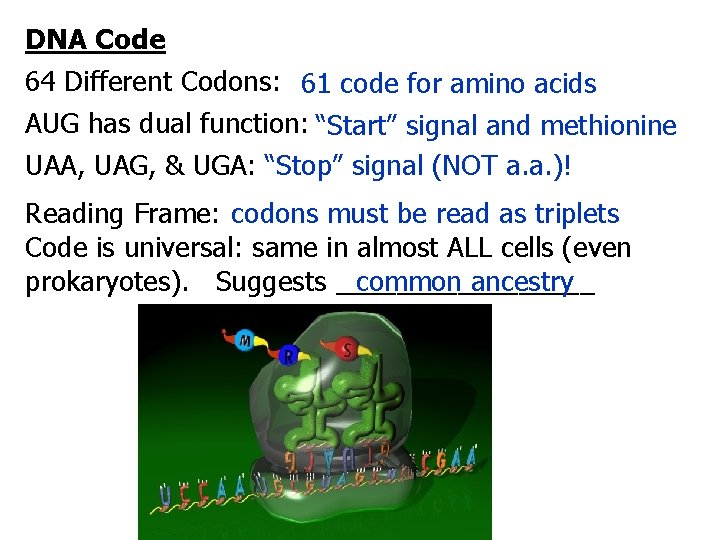
DNA Code 64 Different Codons: 61 code for amino acids AUG has dual function: “Start” signal and methionine UAA, UAG, & UGA: “Stop” signal (NOT a. a. )! Reading Frame: codons must be read as triplets Code is universal: same in almost ALL cells (even common ancestry prokaryotes). Suggests _________
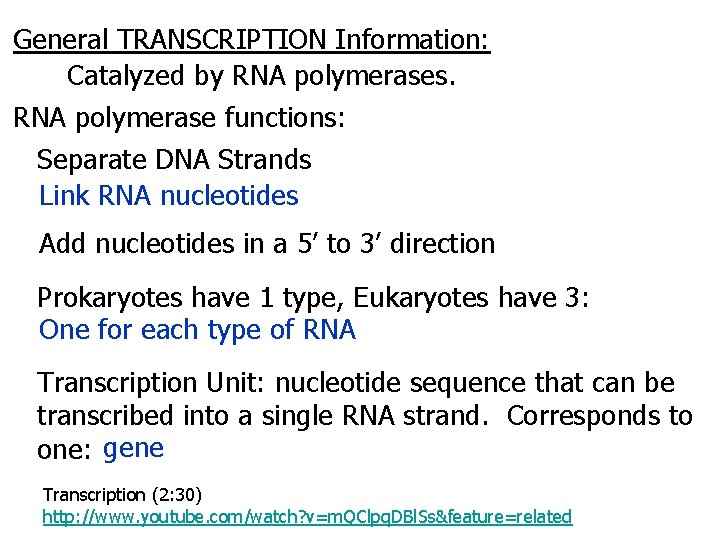
General TRANSCRIPTION Information: Catalyzed by RNA polymerases. RNA polymerase functions: Separate DNA Strands Link RNA nucleotides Add nucleotides in a 5’ to 3’ direction Prokaryotes have 1 type, Eukaryotes have 3: One for each type of RNA Transcription Unit: nucleotide sequence that can be transcribed into a single RNA strand. Corresponds to one: gene Transcription (2: 30) http: //www. youtube. com/watch? v=m. QClpq. DBl. Ss&feature=related
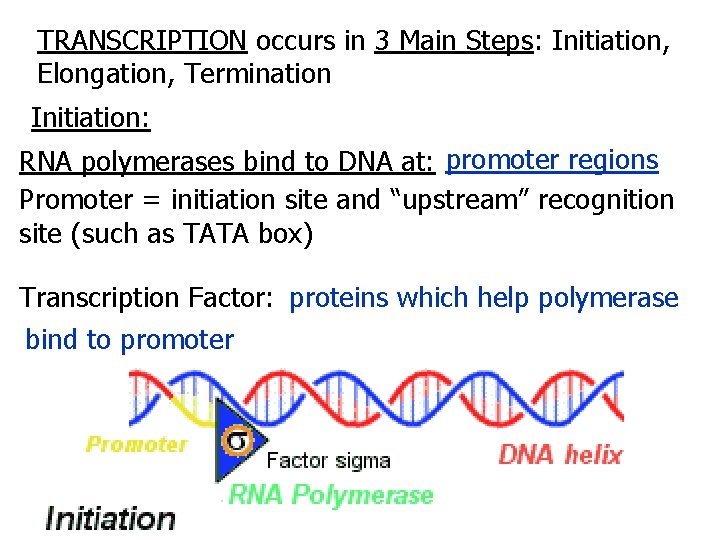
TRANSCRIPTION occurs in 3 Main Steps: Initiation, Elongation, Termination Initiation: RNA polymerases bind to DNA at: promoter regions Promoter = initiation site and “upstream” recognition site (such as TATA box) Transcription Factor: proteins which help polymerase bind to promoter
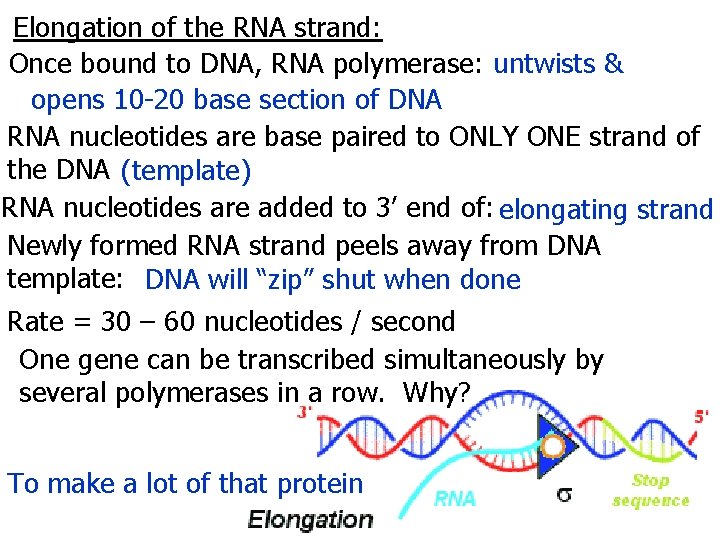
Elongation of the RNA strand: Once bound to DNA, RNA polymerase: untwists & opens 10 -20 base section of DNA RNA nucleotides are base paired to ONLY ONE strand of the DNA (template) RNA nucleotides are added to 3’ end of: elongating strand Newly formed RNA strand peels away from DNA template: DNA will “zip” shut when done Rate = 30 – 60 nucleotides / second One gene can be transcribed simultaneously by several polymerases in a row. Why? To make a lot of that protein
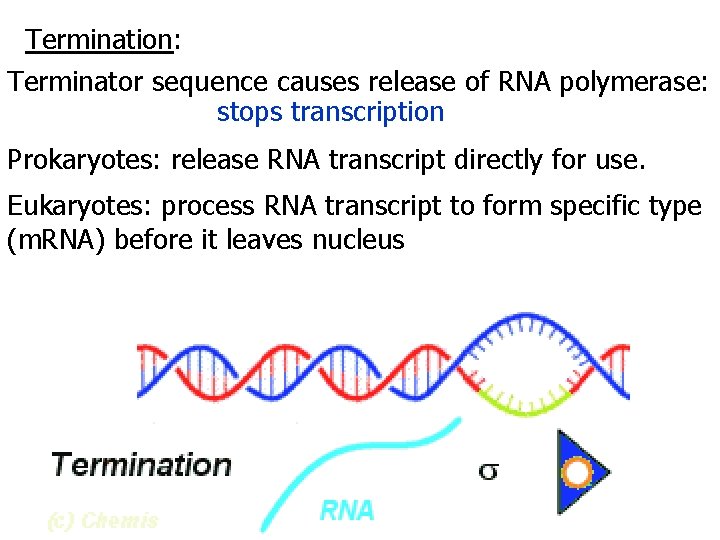
Termination: Terminator sequence causes release of RNA polymerase: stops transcription Prokaryotes: release RNA transcript directly for use. Eukaryotes: process RNA transcript to form specific type (m. RNA) before it leaves nucleus
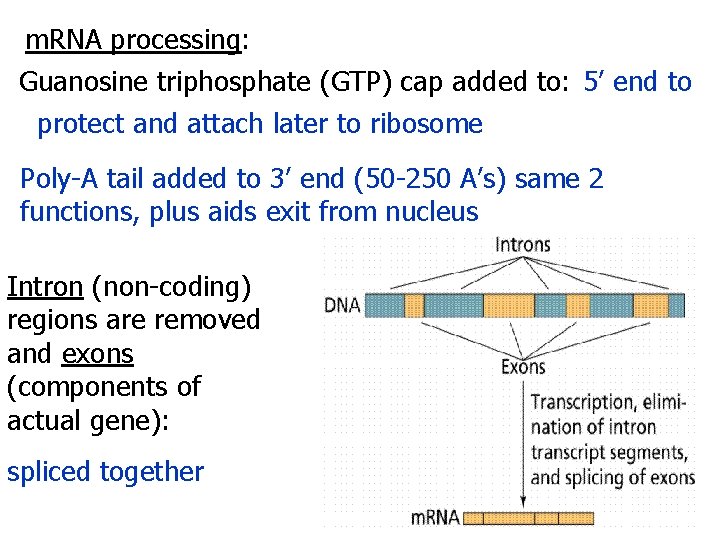
m. RNA processing: Guanosine triphosphate (GTP) cap added to: 5’ end to protect and attach later to ribosome Poly-A tail added to 3’ end (50 -250 A’s) same 2 functions, plus aids exit from nucleus Intron (non-coding) regions are removed and exons (components of actual gene): spliced together

Functions of Introns? Possible Regulatory Role: gene activity, possibly helps m. RNA exit nucleus Allows one gene to have multiple functions: alternative RNA splicing gives rise to diff. protein Evolutionary Role: Allows for increased chance of crossing over within a gene: produces novel versions of a gene. Increases recombination.
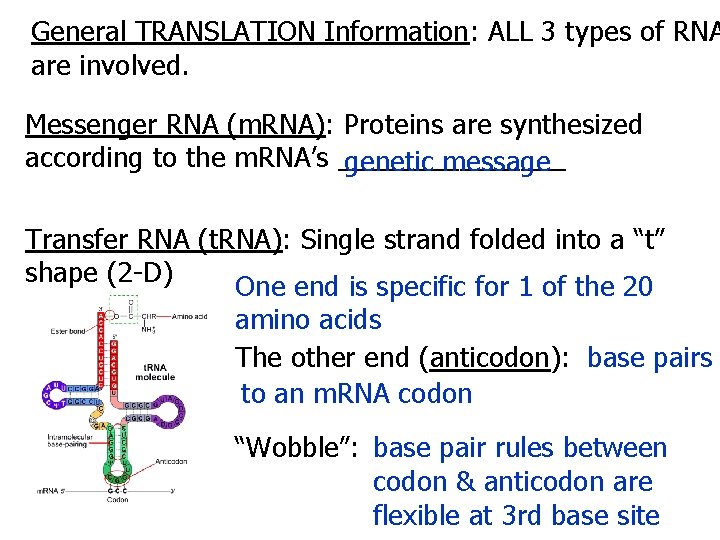
General TRANSLATION Information: ALL 3 types of RNA are involved. Messenger RNA (m. RNA): Proteins are synthesized according to the m. RNA’s ________ genetic message Transfer RNA (t. RNA): Single strand folded into a “t” shape (2 -D) One end is specific for 1 of the 20 amino acids The other end (anticodon): base pairs to an m. RNA codon “Wobble”: base pair rules between codon & anticodon are flexible at 3 rd base site
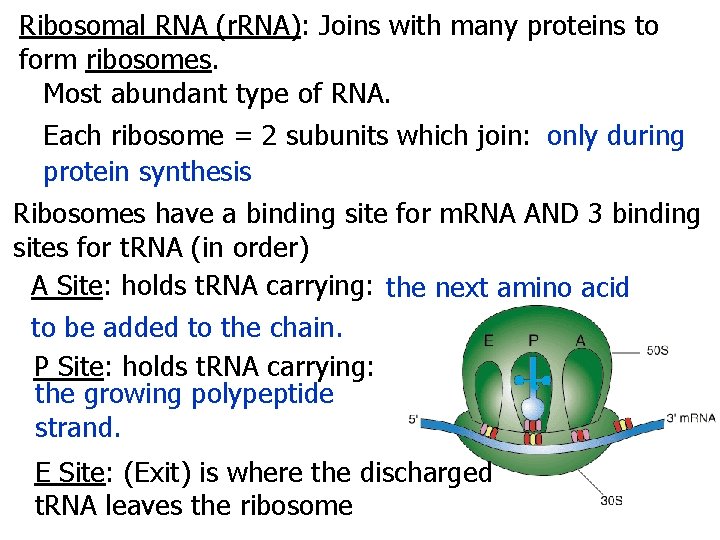
Ribosomal RNA (r. RNA): Joins with many proteins to form ribosomes. Most abundant type of RNA. Each ribosome = 2 subunits which join: only during protein synthesis Ribosomes have a binding site for m. RNA AND 3 binding sites for t. RNA (in order) A Site: holds t. RNA carrying: the next amino acid to be added to the chain. P Site: holds t. RNA carrying: the growing polypeptide strand. E Site: (Exit) is where the discharged t. RNA leaves the ribosome
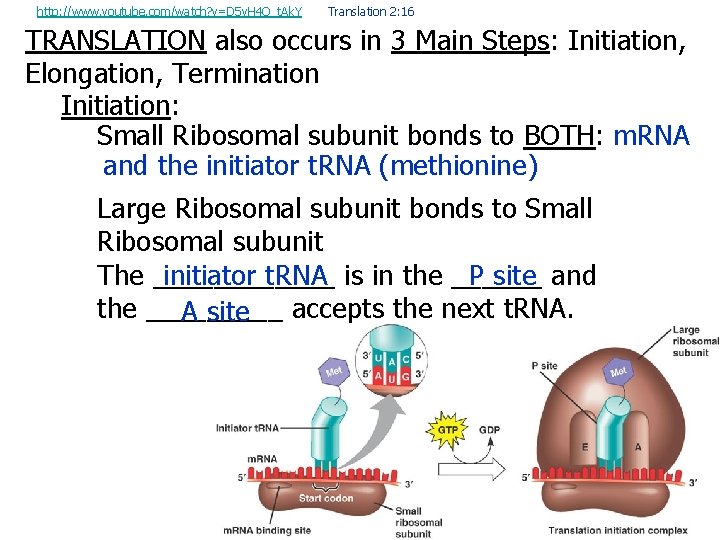
http: //www. youtube. com/watch? v=D 5 v. H 4 Q_t. Ak. Y Translation 2: 16 TRANSLATION also occurs in 3 Main Steps: Initiation, Elongation, Termination Initiation: Small Ribosomal subunit bonds to BOTH: m. RNA and the initiator t. RNA (methionine) Large Ribosomal subunit bonds to Small Ribosomal subunit initiator t. RNA is in the ______ P site and The ______ the _____ accepts the next t. RNA. A site
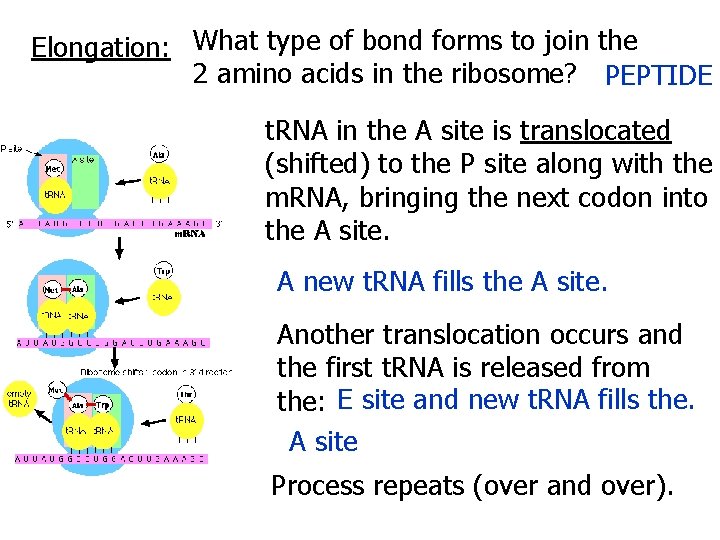
Elongation: What type of bond forms to join the 2 amino acids in the ribosome? PEPTIDE t. RNA in the A site is translocated (shifted) to the P site along with the m. RNA, bringing the next codon into the A site. A new t. RNA fills the A site. Another translocation occurs and the first t. RNA is released from the: E site and new t. RNA fills the. A site Process repeats (over and over).
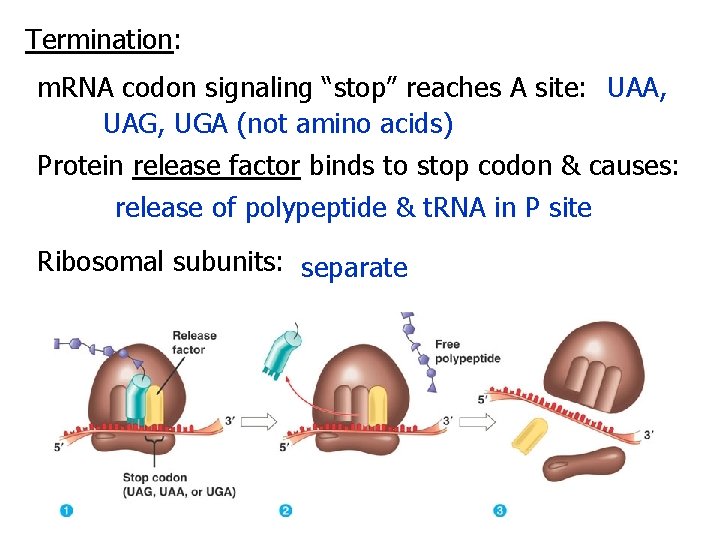
Termination: m. RNA codon signaling “stop” reaches A site: UAA, UAG, UGA (not amino acids) Protein release factor binds to stop codon & causes: release of polypeptide & t. RNA in P site Ribosomal subunits: separate
MUTATIONS & their effects on Proteins Mutation = a permanent change in DNA Point mutations = change in 1 nucleotide pair
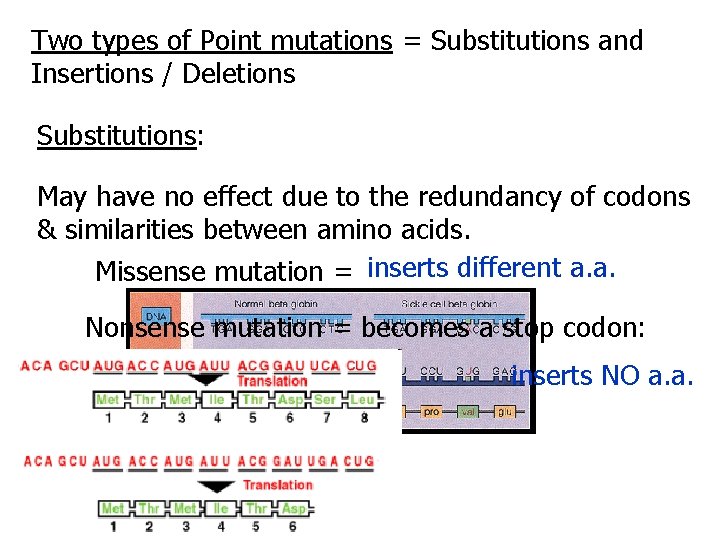
Two types of Point mutations = Substitutions and Insertions / Deletions Substitutions: May have no effect due to the redundancy of codons & similarities between amino acids. Missense mutation = inserts different a. a. Nonsense mutation = becomes a stop codon: inserts NO a. a.
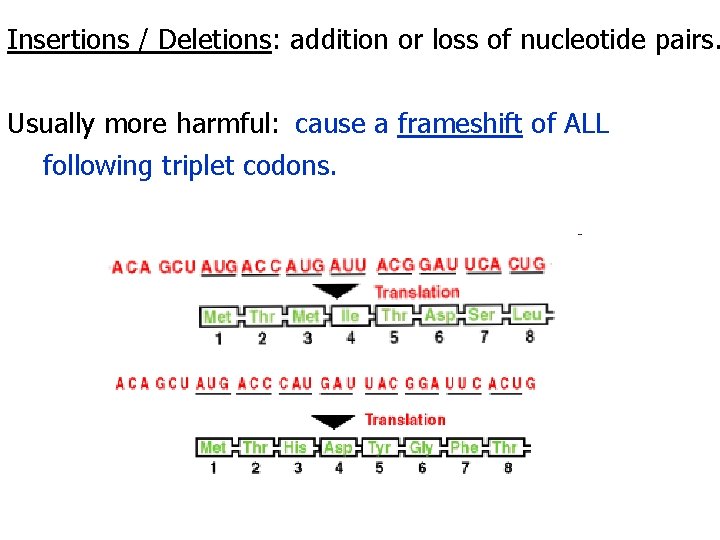
Insertions / Deletions: addition or loss of nucleotide pairs. Usually more harmful: cause a frameshift of ALL following triplet codons.
Mutagenesis= the creation of mutations Spontaneous: mistakes in DNA replication, repair, or recombination Exposure to mutagens (physical or chemical agents): radiation, drugs. . . (many are carcinogens)
- Slides: 40