Highdensity admixture mapping to find genes for complex

















- Slides: 31

High-density admixture mapping to find genes for complex disease David Reich Harvard Medical School Department of Genetics Broad Institute July 13, 2004 (work with Nick Patterson)

Why do we want to find disease-causing variants? • Identify new targets for rational drug design and treatment • Identify new biological pathways • Clinical genetic testing

Linkage mapping doesn’t work well for common diseases Breast cancer Diabetes Heart attack Manic depression Obesity High cholesterol Multiple sclerosis High blood pressure Stroke Turn to association methods instead

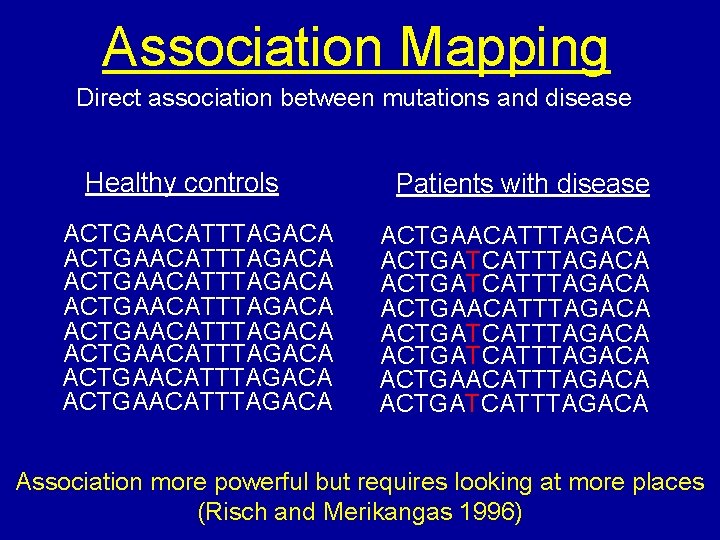
Association Mapping Direct association between mutations and disease Healthy controls ACTGAACATTTAGACA ACTGAACATTTAGACA Patients with disease ACTGAACATTTAGACA ACTGATCATTTAGACA ACTGAACATTTAGACA ACTGATCATTTAGACA Association more powerful but requires looking at more places (Risch and Merikangas 1996)

Admixture mapping. In favorable circumstances, the most economical method for a whole-genome scan 1) The idea of admixture mapping 2) Methods 3) A practical whole-genome map 4) Two real studies

Admixture Mapping (type of association mapping) • Can be as powerful as haplotype association but requires 100 - to 500 -times fewer SNPs • Populations like African and Hispanic Americans • Most promising for diseases with different population risks: multiple sclerosis, prostate cancer, …

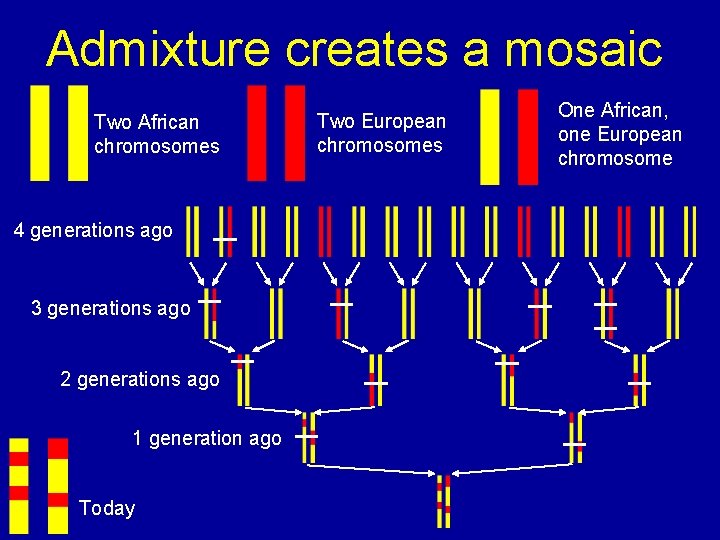
Admixture creates a mosaic Two African chromosomes 4 generations ago 3 generations ago 2 generations ago 1 generation ago Today Two European chromosomes One African, one European chromosome

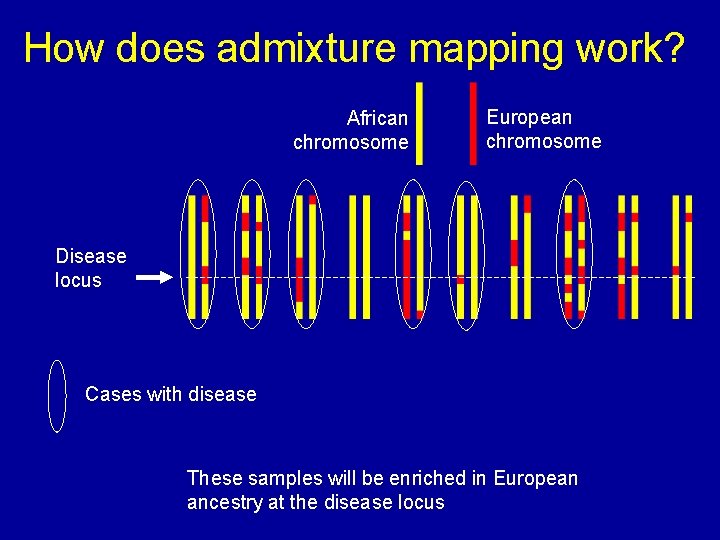
How does admixture mapping work? African chromosome European chromosome Disease locus Cases with disease These samples will be enriched in European ancestry at the disease locus

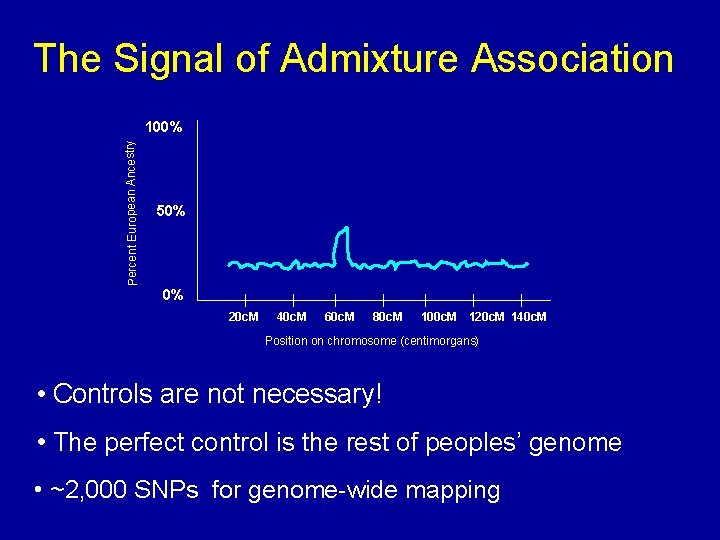
The Signal of Admixture Association Percent European Ancestry 100% 50% 0% 20 c. M 40 c. M 60 c. M 80 c. M 100 c. M 120 c. M 140 c. M Position on chromosome (centimorgans) • Controls are not necessary! • The perfect control is the rest of peoples’ genome • ~2, 000 SNPs for genome-wide mapping

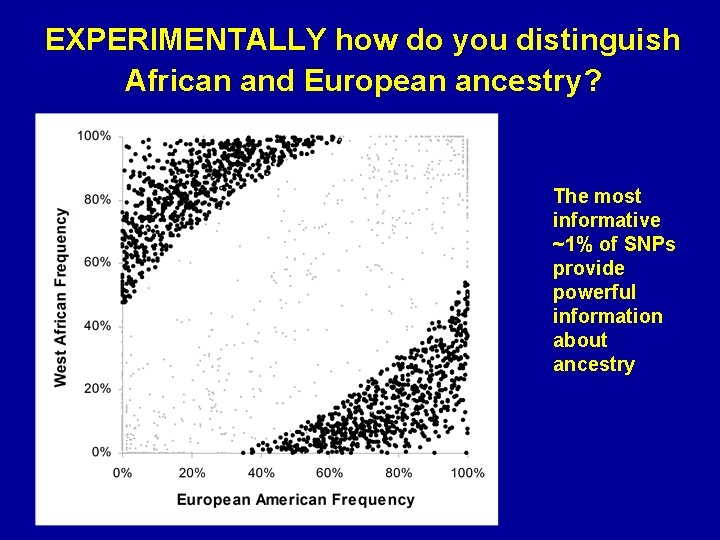
EXPERIMENTALLY how do you distinguish African and European ancestry? The most informative ~1% of SNPs provide powerful information about ancestry

How does one identify European or African segments despite similar gene frequencies? African American Europeans West Africans Strong evidence of African ancestry No evidence of one ancestry or another Strong evidence of European ancestry 100 kb

New Methods • The Hidden Markov Model (for combining information from closely linked, partially informative markers to make inferences about ancestry) • The Markov Chain Monte Carlo (to deal with uncertainties in the HMM parameters that can produce false-positives in analysis)

How to track regions of European & African ancestry along the genome? Mi = % European ancestry in individual’s ancestors >40 generations ago li = Number of generations since mixture Key parameters for the HMM

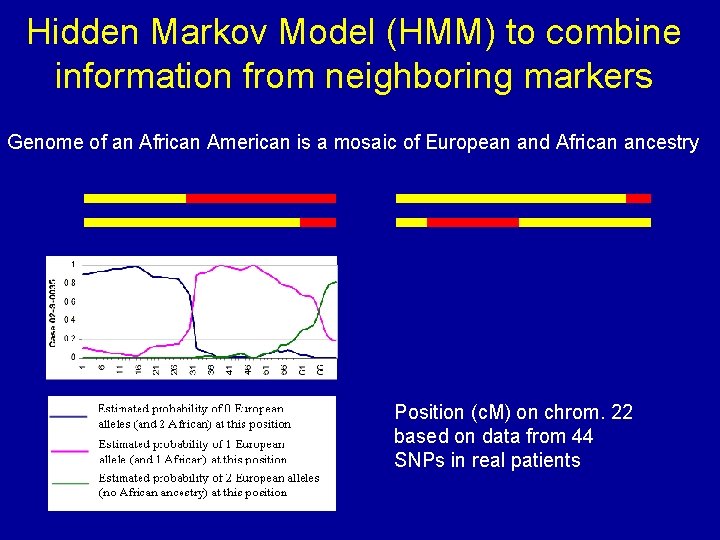
Hidden Markov Model (HMM) to combine information from neighboring markers Genome of an African American is a mosaic of European and African ancestry Position (c. M) on chrom. 22 based on data from 44 SNPs in real patients


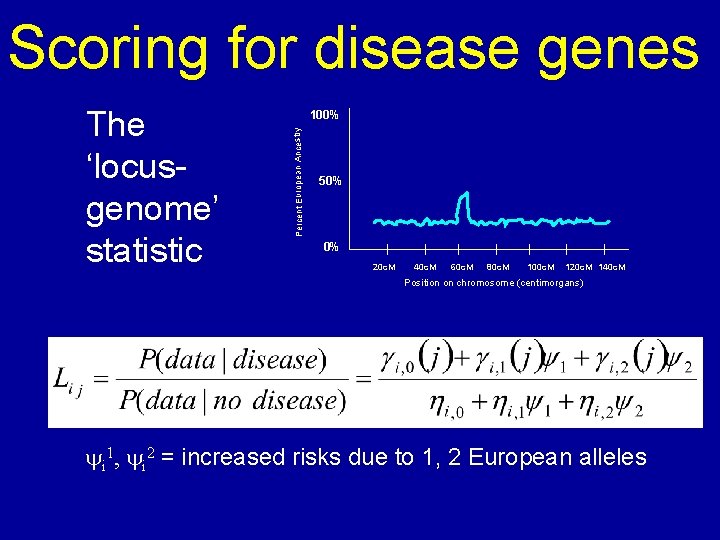
Scoring for disease genes Percent European Ancestry The ‘locusgenome’ statistic 100% 50% 0% 20 c. M 40 c. M 60 c. M 80 c. M 100 c. M 120 c. M 140 c. M Position on chromosome (centimorgans) 2 2 hi, 1 risks = 2 M ) 2 European hi, 2 = (1 -M yhi 1 i, 0 , y=i 2 M = iincreased due to i 1, alleles i(1 -M i)

Can detect regions of increased European ancestry in a data set of 756 SNPs and 442 samples Section 3

Problem with the HMM pj. European and pj. African are assumed known In fact, they are unknown due to… • sampling error when genotyping the parental populations • modern populations aren’t the true parental populations This can cause false-positives!

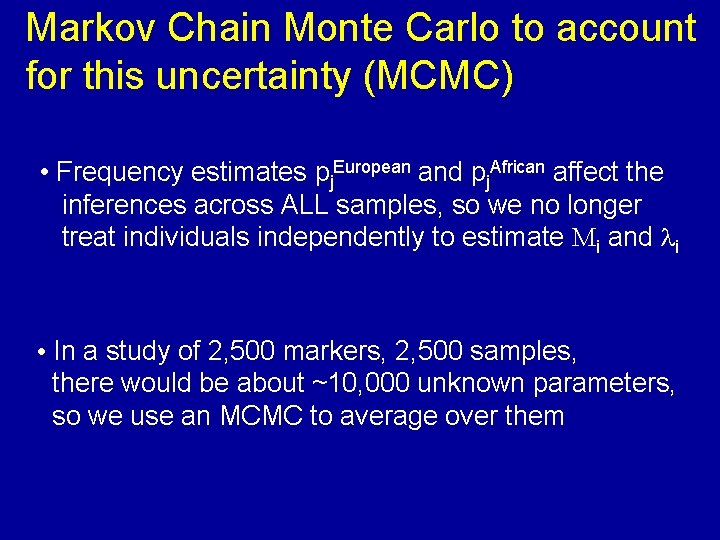
Markov Chain Monte Carlo to account for this uncertainty (MCMC) • Frequency estimates pj. European and pj. African affect the inferences across ALL samples, so we no longer treat individuals independently to estimate Mi and li • In a study of 2, 500 markers, 2, 500 samples, there would be about ~10, 000 unknown parameters, so we use an MCMC to average over them

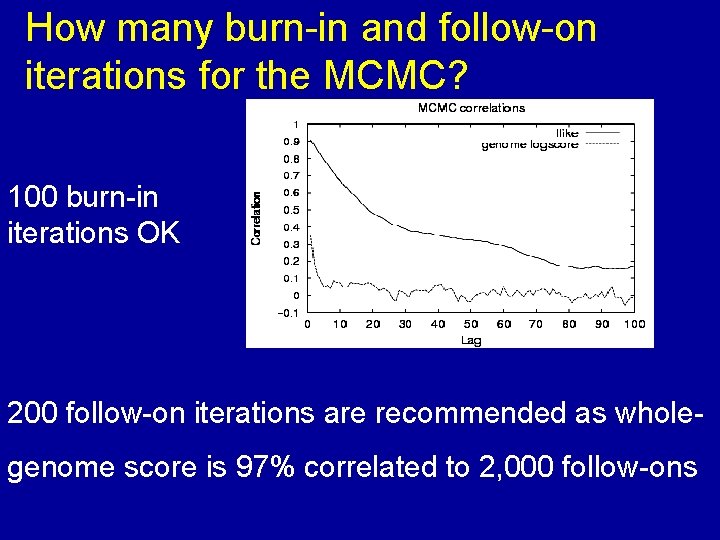
How many burn-in and follow-on iterations for the MCMC? 100 burn-in iterations OK 200 follow-on iterations are recommended as wholegenome score is 97% correlated to 2, 000 follow-ons

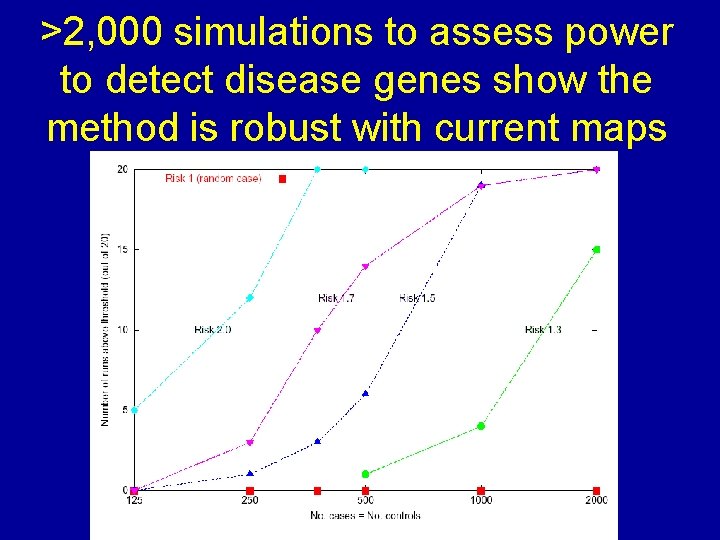
>2, 000 simulations to assess power to detect disease genes show the method is robust with current maps

Genotypes required for whole-genome scans with admixture, linkage and haplotype mapping Risk = 1. 5 Risk = 2. 0 50% allele in Africans 5% allele in Africans Risk = 1. 3 Europeanfrequency (at 80% power) Europeanfrequency

Making admixture mapping work 1) >2, 000 samples required for a powerful study (far more than the ~300 previously recommended) (no controls strictly necessary – cases from one study are controls for another) 2) Diseases to study Hypertension, End-stage renal disease, prostate cancer, Multiple sclerosis, ovarian cancer, Alzheimer’s disease, Type II diabetes (Hispanic Americans) Note: 10 -30% more samples to study diseases prevalent in Africans 3) New resources · High density 2, 154 marker map, 50 x more powerful than before · Powerful, conservative methods (ANCESTRYMAP program)

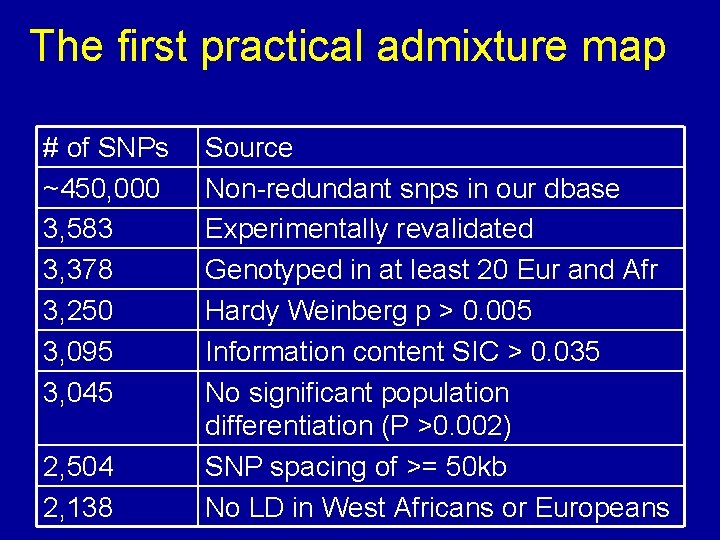
The first practical admixture map # of SNPs ~450, 000 3, 583 3, 378 3, 250 3, 095 3, 045 2, 504 2, 138 Source Non-redundant snps in our dbase Experimentally revalidated Genotyped in at least 20 Eur and Afr Hardy Weinberg p > 0. 005 Information content SIC > 0. 035 No significant population differentiation (P >0. 002) SNP spacing of >= 50 kb No LD in West Africans or Europeans


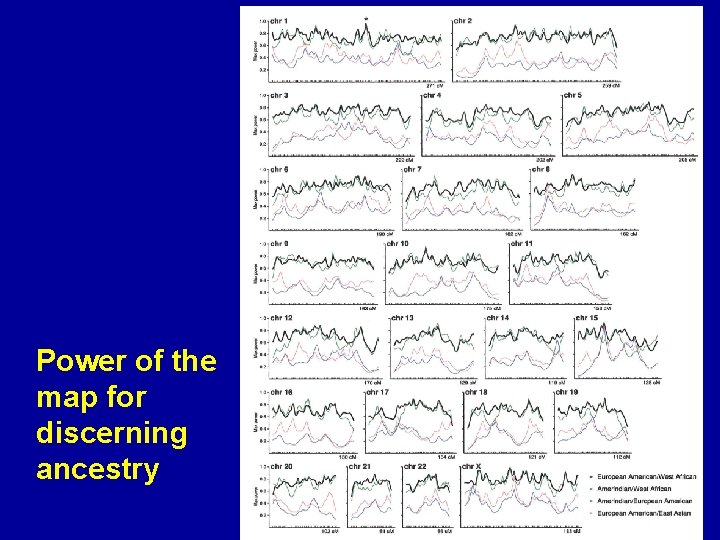
Power of the map for discerning ancestry

Our first two large-scans • Prostate cancer 2 -3 fold more prevalent in African Americans 650 cases, 698 controls already in lab • Multiple sclerosis 1. 5 -2 fold more prevalent in European Americans 502 cases, 175 controls already in lab

Initial screen of 39% of the genome focusing on linkage peaks in 442 MS patients Nothing compelling yet

Currently planning to increase power Targeted power Current multiple sclerosis data ‘theoretically’ 50% power loss due to current map

Conclusions • The imperative now is on finding something with this new method • Must do SEVERAL large-scale studies to assess whether admixture mapping works

Acknowledgements Methods New map Nick Patterson Mike Smith Steve O’Brien Dennis Gilbert Francisco de la Vega Trevor Woodage Charles Scafe Nick Patterson Gavin Mc. Donald Alicja Walizewska David Altshuler Neil Hattangadi Multiple sclerosis David Hafler Nick Patterson Gavin Mc. Donald Alicja Waliszewska Phil de Jager Jorge Oksenberg Stephen Hauser Amy Swerdlin Bruce Cree Robin Lincoln Cari de Loa Prostate caner Matt Freedman David Altshuler Chris Haiman Brian Henderson