High Performance Computing on Flux EEB 401 Charles

High Performance Computing on Flux EEB 401 Charles J Antonelli Mark Champe LSAIT ARS September, 2014
Flux is a university-wide shared computational discovery / high-performance computing service. Provided by Advanced Research Computing at U-M Operated by CAEN HPC Procurement, licensing, billing by U-M ITS Interdisciplinary since 2010 http: //arc. research. umich. edu/resources-services/flux/ cja 2014 2 9/14
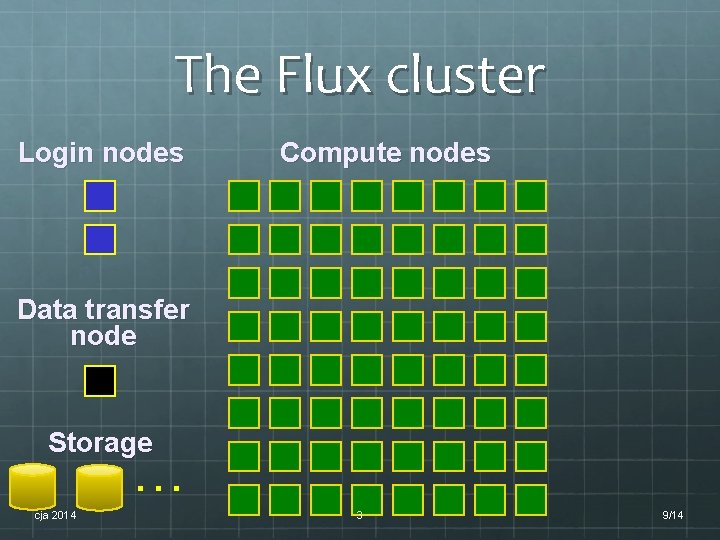
The Flux cluster Login nodes Compute nodes Data transfer node Storage … cja 2014 3 9/14
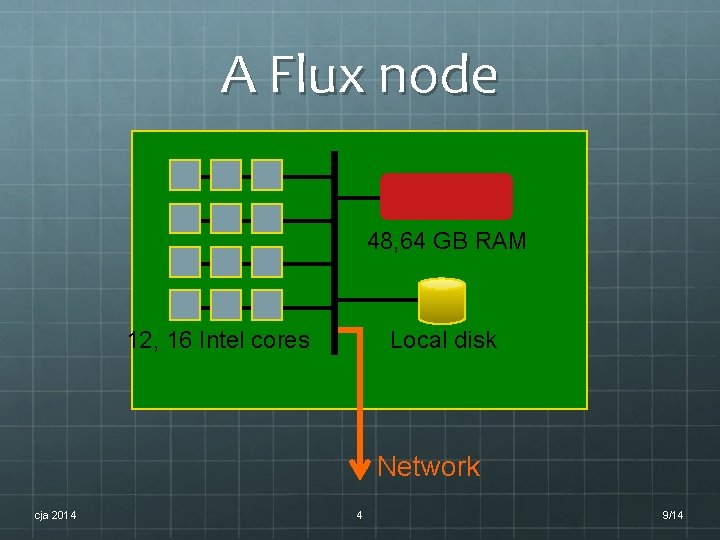
A Flux node 48, 64 GB RAM 12, 16 Intel cores Local disk Network cja 2014 4 9/14
Programming Models Two basic parallel programming models Message-passing The application consists of several processes running on different nodes and communicating with each other over the network Used when the data are too large to fit on a single node, and simple synchronization is adequate “Coarse parallelism” Implemented using MPI (Message Passing Interface) libraries Multi-threaded The application consists of a single process containing several parallel threads that communicate with each other using synchronization primitives Used when the data can fit into a single process, and the communications overhead of the message-passing model is intolerable “Fine-grained parallelism” or “shared-memory parallelism” Implemented using Open. MP (Open Multi-Processing) compilers and libraries Both cja 2014 5 9/14

Command Line Reference William E Shotts, Jr. , “The Linux Command Line: A Complete Introduction, ” No Starch Press, January 2012. http: //linuxcommand. org/tlcl. php. Download Creative Commons Licensed version athttp: //downloads. sourceforge. net/project/linuxcomma nd/TLCL/13. 07/TLCL-13. 07. pdf. cja 2014 6 9/14

Using Flux Three basic requirements: A Flux login account A Flux allocation An MToken (or a Software Token) Logging in to Flux ssh login@flux-login. engin. umich. edu Campus wired or MWireless VPN ssh login. itd. umich. edu first cja 2014 7 9/14
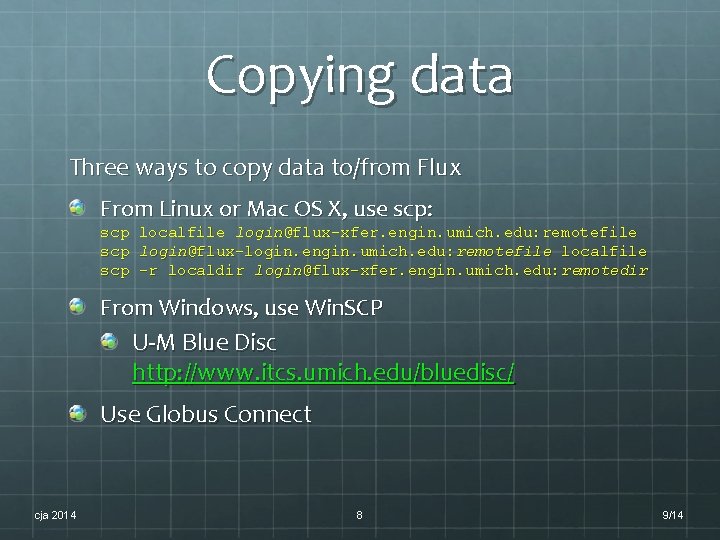
Copying data Three ways to copy data to/from Flux From Linux or Mac OS X, use scp: scp scp localfile login@flux-xfer. engin. umich. edu: remotefile login@flux-login. engin. umich. edu: remotefile localfile -r localdir login@flux-xfer. engin. umich. edu: remotedir From Windows, use Win. SCP U-M Blue Disc http: //www. itcs. umich. edu/bluedisc/ Use Globus Connect cja 2014 8 9/14

Globus Online Features High-speed data transfer, much faster than scp or Win. SCP Reliable & persistent Minimal client software: Mac OS X, Linux, Windows Grid. FTP Endpoints Gateways through which data flow Exist for XSEDE, OSG, … UMich: umich#flux, umich#nyx Add your own client endpoint! Add your own server endpoint: contact flux-support@umich. edu More information http: //cac. engin. umich. edu/resources/login-nodes/globus- gridftp cja 2014 9 9/14

Batch workflow You create a batch script and submit it to PBS (the cluster resource manager & scheduler) PBS schedules your job, and it enters the flux queue When its turn arrives, your job will execute the batch script Your script has access to any applications or data stored on the Flux cluster When your job completes, anything it sent to standard output and error are saved and returned to you You can check on the status of your job at any time, or delete it if it’s not doing what you want A short time after your job completes, it disappears cja 2014 10 9/14
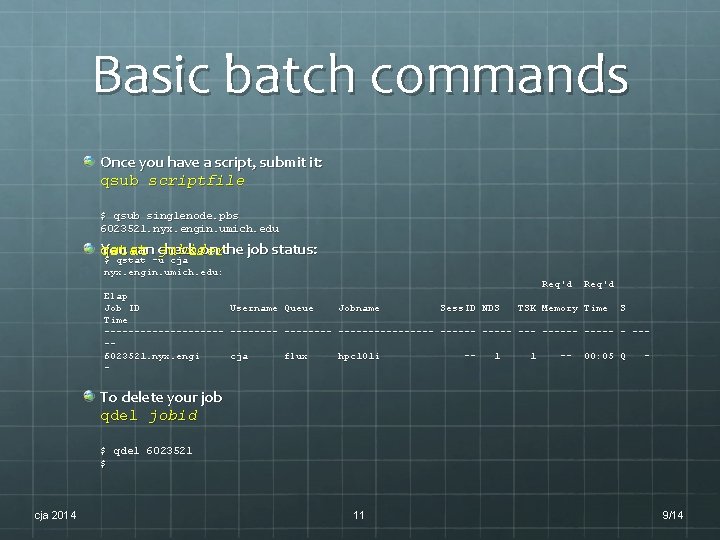
Basic batch commands Once you have a script, submit it: qsub scriptfile $ qsub singlenode. pbs 6023521. nyx. engin. umich. edu You can check on the job status: jobid qstat -u user $ qstat -u cja nyx. engin. umich. edu: Req'd Elap Job ID Username Queue Jobname Sess. ID NDS TSK Memory Time S Time ---------- -------- --- ----- - ---6023521. nyx. engi cja flux hpc 101 i -1 1 -- 00: 05 Q -- To delete your job qdel jobid $ qdel 6023521 $ cja 2014 11 9/14

Loosely-coupled batch script #PBS -N yourjobname #PBS -V #PBS -A youralloc_flux #PBS -l qos=flux #PBS -q flux #PBS –l procs=12, pmem=1 gb, walltime=01: 00 #PBS -M youremailaddress #PBS -m abe #PBS -j oe #Your Code Goes Below: cd $PBS_O_WORKDIR mpirun. /c_ex 01 cja 2014 12 9/14

Tightly-coupled batch script #PBS -N yourjobname #PBS -V #PBS -A youralloc_flux #PBS -l qos=flux #PBS -q flux #PBS –l nodes=1: ppn=12, mem=47 gb, walltime=02: 00 #PBS -M youremailaddress #PBS -m abe #PBS -j oe #Your Code Goes Below: cd $PBS_O_WORKDIR matlab -nodisplay -r script cja 2014 13 9/14

Flux software Licensed and open software: Abacus, BLAST, BWA, bowtie, ANSYS, Java, Mason, Mathematica, Matlab, R, RSEM, STATA SE, … See http: //cac. engin. umich. edu/resources C, C++, Fortran compilers: Intel (default), PGI, GNU toolchains You can choose software using the module command cja 2014 14 9/14

Modules The module command allows you to specify what versions of software you want to use module module list -- Show loaded modules load name -- Load module name for use show name -- Show info for name avail -- Show all available modules avail name -- Show versions of module name* unload name -- Unload module name -- List all options Enter these commands at any time during your session A configuration file allows default module commands to be executed at login Put module commands in file ~/privatemodules/default Don’t put module commands in your. bashrc /. bash_profile cja 2014 15 9/14

Flux storage Lustre filesystem mounted on /scratch on all login, compute, and transfer nodes 640 TB of short-term storage for batch jobs Large, fast, short-term NFS filesystems mounted on /home and /home 2 on all nodes 80 GB of storage per user for development & testing Small, slow, long-term cja 2014 16 9/14

Flux environment The Flux login nodes have the standard GNU/Linux toolkit: make, perl, python, java, emacs, vi, nano, … Watch out for source code or data files written on non-Linux systems Use these tools to analyze and convert source files to Linux format file dos 2 unix cja 2014 17 9/14

BLAST Load modules mod unload intel-comp openmpi gcc mod load med python/3. 2. 3 gcc boost/1. 54. 0 -gcc ncbi-blast/2. 2. 29 Create file ~/. ncbirc , with contents [BLAST] BLASTDB=/nfs/med-ref-genomes/blast Copy sample code to your home directory cd cp ~cja/hpc/eeb 401 -sample-code. tar. gz. tar -zxvf eeb 401 -sample-code. tar. gz cd. /eeb 401 -sample-code cja 2014 18 9/14
BLAST Examine blast-example. pbs Edit with your favorite Linux editor emacs, vi, pico, … Change email address bjensen@umich. edu to your own cja 2014 19 9/14
BLAST Submit your job to Flux qsub blast-example. pbs Watch the progress of your job qstat jobid When complete, look at the job’s output less blast-example. ojobid cja 2014 20 9/14

BWA module cja 2014 load med med samtools ncbi-blast bowtie # optional bwa 21 9/14

Bowtie module load med bowtie cja 2014 22 9/14

RSEM module load R/3. 0. 1 module load lsa rsem module load med bowtie Note: loading R/3. 0. 1 unloads gcc/4. 7. 0 and loads gcc/4. 4. 6 cja 2014 23 9/14

Perl scripts module load lsa baucom-bioinformatics module show baucom-bioinformatics cja 2014 24 9/14

Interactive jobs You can submit jobs interactively: qsub -I -X -V -l procs=2 -l walltime=15: 00 -A youralloc_flux -l qos=flux –q flux This queues a job as usual Your terminal session will be blocked until the job runs When your job runs, you'll get an interactive shell on one of your nodes Invoked commands will have access to all of your nodes When you exit the shell your job is deleted Interactive jobs allow you to Develop and test on cluster node(s) Execute GUI tools on a cluster node Utilize a parallel debugger interactively cja 2014 25 9/14

Interactive BLAST Load modules: module unload gcc openmpi module load med gcc ncbi-blast Start an interactive PBS session qsub -I -V -l nodes=1: ppn=2 -l walltime=1: 00 -A eeb 401 f 14_flux -l qos=flux -q flux Run BLAST in the interactive shell cd $PBS_O_WORKDIR blastdbcmd -db refseq_rna -entry nm_000249 -out test_query. fa blastn -query test_query. fa -db refseq_rna -task blastn -dust no -outfmt 7 -num_alignments 2 -num_descriptions 2 -num_threads 2 cja 2014 26 9/14

Gaining insight There are several commands you can run to get some insight over when your job will start: freenodes : shows the total number of free nodes and cores currently available on Flux mdiag -a youralloc_name : shows cores and memory defined for your allocation and who can run against it showq -w acct=yourallocname: shows cores being used by jobs running against your allocation (running/idle/blocked) checkjob -v jobid : Can show why your job might not be starting showstart -e all jobid : Gives you a coarse estimate of job start time; use the smallest value returned cja 2014 27 9/14

Some Flux Resources http: //arc. research. umich. edu/resources-services/flux/ U-M Advanced Research Computing Flux pages http: //cac. engin. umich. edu/ CAEN HPC Flux pages http: //www. youtube. com/user/UMCo. ECAC CAEN HPC You. Tube channel For assistance: hpc-support@umich. edu Read by a team of people including unit support staff Cannot help with programming questions, but can help with operational Flux and basic usage questions cja 2014 28 9/14

Any Questions? Charles J. Antonelli LSAIT Advocacy and Research Support cja@umich. edu http: //www. umich. edu/~cja 734 763 0607 cja 2014 29 9/14
- Slides: 29