Hidden Markov Models That Use Predicted Local Structure




- Slides: 16

Hidden Markov Models That Use Predicted Local Structure for Fold Recognition: Alphabets of Backbone Geometry R Karchin, M Cline, Y Mandel. Gutfreund, K Karplus

Problem • Parent fold may have low sequence similarity • Most often, query is sequence-only

Proposed Solution • Try to predict local structure of target • Use local structure prediction in fold recognition

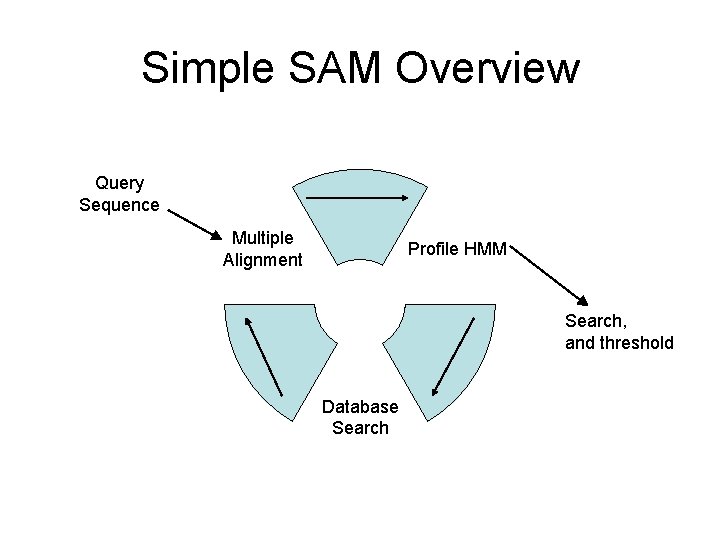
Simple SAM Overview Query Sequence Multiple Alignment Profile HMM Search, and threshold Database Search

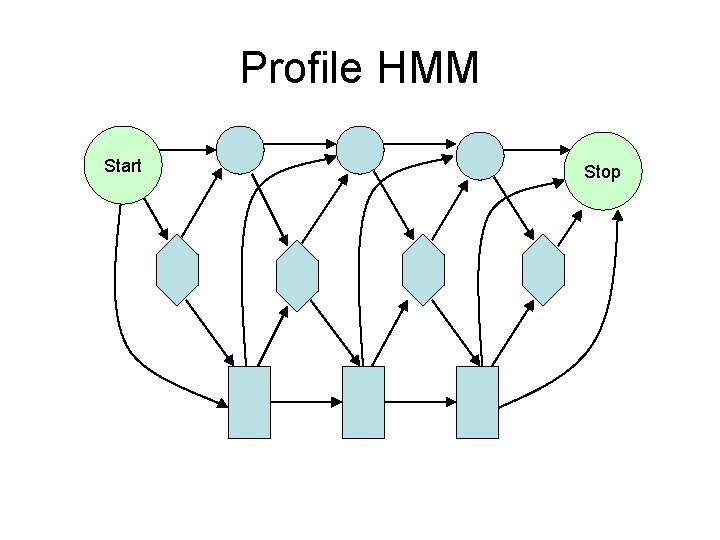
Profile HMM Start Stop

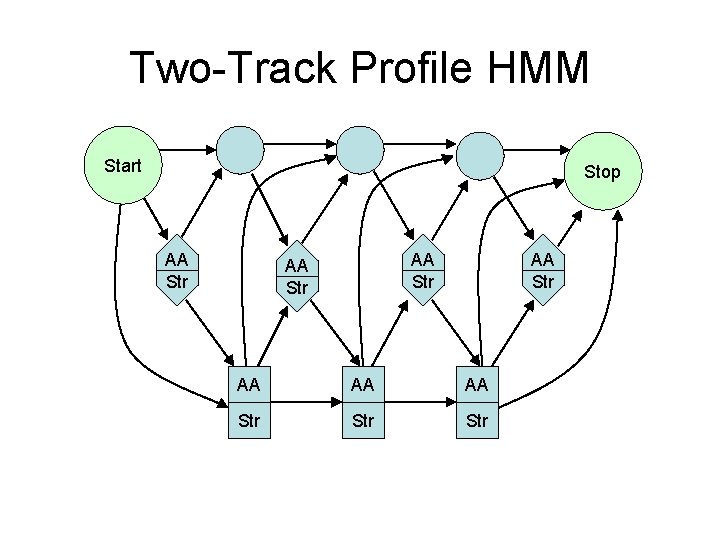
Two-Track Profile HMM Start Stop AA Str AA AA AA Str Str

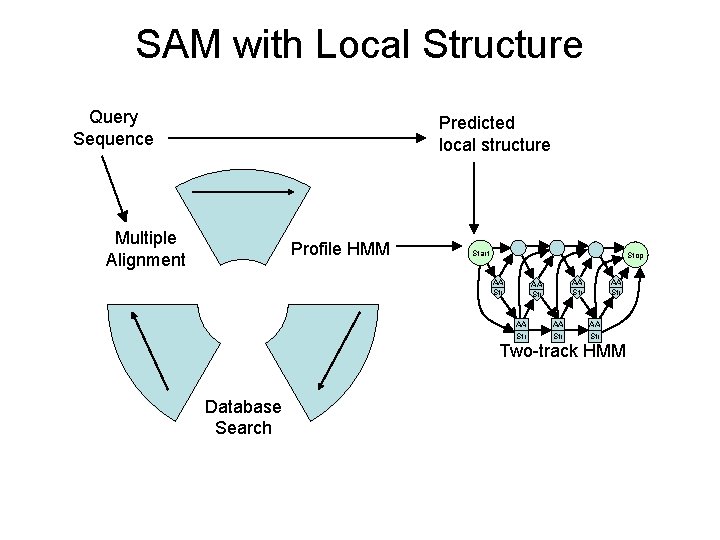
SAM with Local Structure Query Sequence Predicted local structure Multiple Alignment Profile HMM Start Stop AA Str AA Str Two-track HMM Database Search

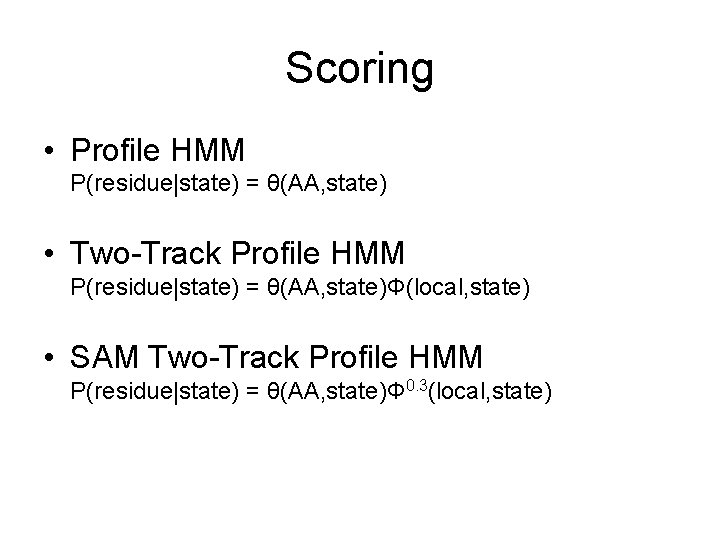
Scoring • Profile HMM P(residue|state) = θ(AA, state) • Two-Track Profile HMM P(residue|state) = θ(AA, state)Φ(local, state) • SAM Two-Track Profile HMM P(residue|state) = θ(AA, state)Φ 0. 3(local, state)

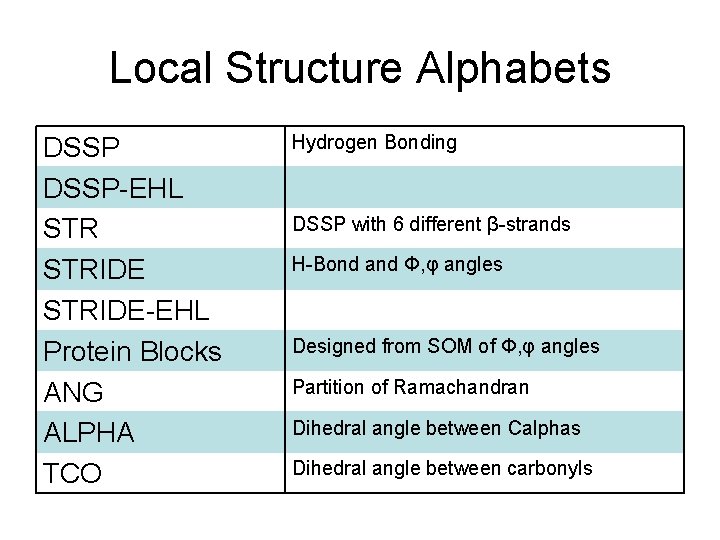
Local Structure Alphabets DSSP-EHL STRIDE-EHL Protein Blocks ANG ALPHA TCO Hydrogen Bonding DSSP with 6 different β-strands H-Bond and Φ, φ angles Designed from SOM of Φ, φ angles Partition of Ramachandran Dihedral angle between Calphas Dihedral angle between carbonyls

Predicting Local Structure of the Query • Alphabets very dissimilar • Use a neural network – Input: window of multiple alignment – Output: structure probabilities for a single residue

Good Structure Alphabets • Intuitively, we want – Predictability – Conservation – Better fold recognition – Better alignment

Predictability • Evaluated on – Exact predictions (QN) – Overlap of structure segments (SOV) – Information gain • Winners: – *-EHL for precision – STR and PB for most information

Conservation • Used FSSP structural alignments • Calculated mutual information between proteins • Used only alignments with low sequence similarity • Winners: – STR, PB

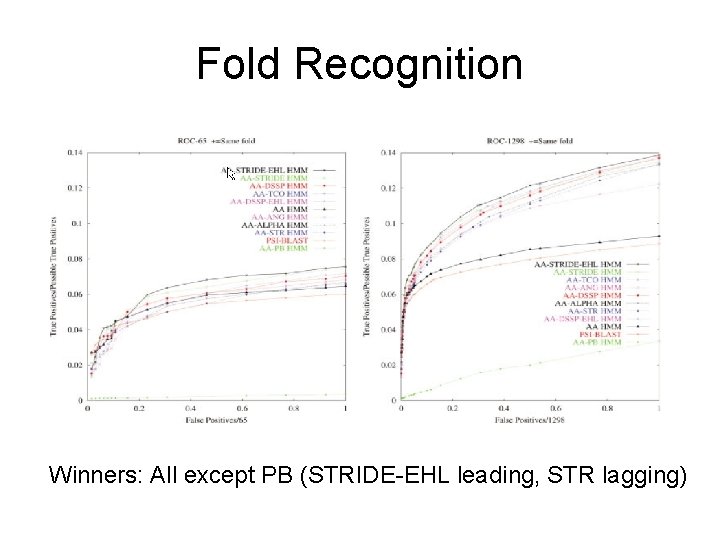
Fold Recognition Winners: All except PB (STRIDE-EHL leading, STR lagging)

Alignment • Compared to DALI and CE alignments • Evaluated using a shift score – 1. 0 perfect match – -0. 2 is the worst (adjustable) • Winners – STR, and STRIDE gets an odd one. – All alphabets improve alignment

Conclusions • Local structure improves: – Fold recognition – Alignment when there’s little sequence similarity • Alignment and fold recognition are very different problems